Figures
Introduction
In developing biologics or biotherapeutics, a great deal of effort is used to ensure that none of the contaminating or hazardous residual components from the organisms responsible for protein expression remain after purification. As a result, additional sanitization steps may be required for the instrumentation utilized in these purification processes.
In this study, microbial challenge was performed on the NGC Chromatography System using a bacterial strain and two yeast strains commonly used in purification environments. These organisms were used to contaminate the entire NGC System for 24 hours, followed by a sanitization process. Sanitization of the system was completed using a cleaning-in-place (CIP) method with 1 M NaOH resulting in a greater than 6 log reduction in microorganisms using colony forming units (CFU) as the test indicator. Extended incubation with 1 M NaOH also reduced endotoxins to acceptable levels. We conclude that the NGC System is capable of being sanitized making it an ideal platform for the small-scale production of biotherapeutics.
Chromatography Instruments in a cGMP Laboratory
Instruments behind chromatography techniques are often considered a vital part of the lab, therefore proper maintenance of the system is crucial.
The Bio-Rad NGC Chromatography System platform is used for purification during drug discovery phases to identify lead candidates, during development phases in which optimization and processes are established, and during transfer of established processes to manufacturing. In a cGMP environment, effective sanitization procedures for small batch production of biopharmaceuticals are required. Validating equipment cleanliness is critical in the pharmaceutical industry and of utmost importance to avoid contaminating drug products with microorganisms, viruses, endotoxins, cleaning solution residues, and the like. Diagnostic tests, such as those used in contaminating an entire chromatography system with living organisms followed by full-system decontamination are important tools to verify the level to which a system can be cleaned.
Sodium hydroxide is an effective antimicrobial agent that can reduce microorganisms to safe levels. It has also been shown to be powerful in denaturing nucleic acids and proteins. Sodium hydroxide concentrations ranging from 0.1 to 1 M with a contact time of 1 to 2 hours is normally sufficient, although endotoxin inactivation may require a contact time of more than 12 hours.
Contaminating the NGC System on Purpose
All modules of a fully equipped NGC platform were contaminated with three different organisms (Escherichia coli, Pichia pastoris, and Saccharomyces cerevisiae) that are typically used for the production of recombinant proteins (Table 1). The focus of these sanitization experiments was cleaning-in-place (CIP) without dismantling the NGC Chromatography System before or after infection. After 24 hours, samples of the infection solutions were taken in order to count the number of surviving organisms. Sampling was followed by decontamination of the NGC platform with 1 M NaOH. Twenty percent ethanol was used to flush out NaOH. Then, the NGC platform was rinsed with a sterile 0.9% NaCl solution to sample for surviving microorganisms, which were subsequently tested using different agar plates.
Acceptance criteria for a successful cleaning process was a minimum 6 log reduction for colony forming units (CFU) and 1 CFU in 10 ml effluent in absolute numbers combined. Endotoxin levels were specifi cally measured after E. coli contamination and cleaning with NaOH. This process demonstrated an effective sanitization.
Table 1. Organisms used for the contamination and sanitization experiments.
| Organism | Escherichia coli | Pichia pastoris | Saccharomyces cerevisiae |
| Specification and selectivity | BL21 (DE3) with vector EX-EGFP-B13 — ampicillin resistance | GS 115 — auxotrophic for histidine | INVSc1 — auxotrophic for histidine, leucine, tryptophan, uracil |
| Growth medium for infection | LBamp | YPD; Minimal dextrose (MD) with specific amino acid | YPD; Minimal dextrose (MD) with specific amino acid |
| Microbial agar testing conditions after decontamination | LB; LBamp; YPD | YPD; MD with and without specific amino acid | YPD; MD with and without specific amino acid |
Details of the Experimental Procedure
Microbial challenge experiments were performed on an NGC Discover 10 Pro Chromatography System (Figure 1). Flow rate was set at 10 ml/min for all experiments. The system contains two types of tubing: PEEK tubing (ID of 0.020″ and 0.030″) and transparent PTFE tubing (ID of 0.062″). Before infection and decontamination, the NGC System was cleaned with 1 M NaOH and 20% ethanol. To detect any remaining background microflora, 20% ethanol was removed with sterile 0.9% NaCl solution and control samples were taken regularly. No CFUs were detected on either YPD or LB agar plates after 24 hours incubation. The back-wash system of the sample pump was also analyzed for background microflora and no CFUs were detected there either.
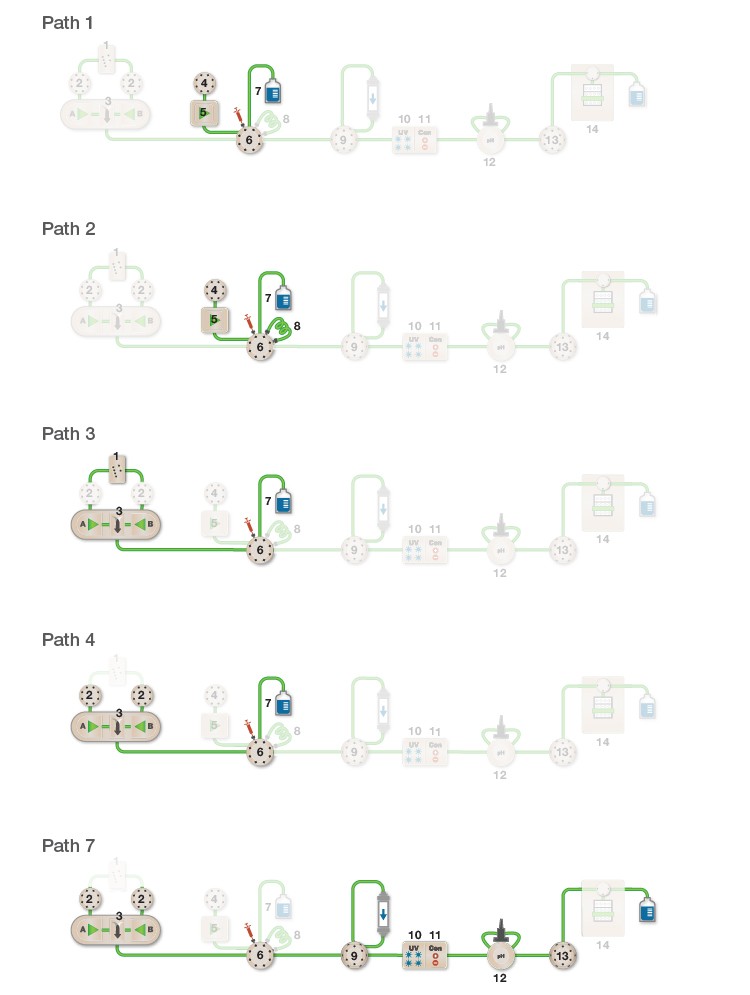
NGC Module Legend
| Module | Module Name |
| 1 | Buffer blending valve |
| 2 | Buffer inlet valves A and B |
| 3 | F10 system pumps A and B with mixer |
| 4 | Sample inlet valve |
| 5 | F100 sample pump |
| 6 | Sample inject valve |
| 7 | Waste position of module 6 |
| 8 | Sample loop (1ml) of module 6 |
| 9 | Column switching valve |
| 10 | Multi-wavelength UV/Vis detector |
| 11 | Conductivity monitor |
| 12 | pH valve |
| 13 | Outlet valve |
| 14 | Fraction collector |
Fig. 1. Sample elution flow paths.
Challenging the NGC System with E. coli-B13
The E. coli-B13 experiments were multifaceted due to the complexity of the system. In the experiments, the NGC System components were infected together, in duplicate, and using multiple flow paths. Samples taken immediately after decontamination showed no signs of growth on LBamp plates. Resampling 24 hours after decontamination indicated no specific bacterial growth. Nonreproducible and unspecific colonies were seen on both YPD and LB agar plates, but greater than a 6 log reduction of bacterial growth was still achieved.
Challenging the NGC System with P. pastoris– GS 115 and S. cerevisiae-INVSc1
The P. pastoris-GS 115 and S. cerevisiae-INVSc1 experiments were performed in duplicate. No viable P. pastoris-GS 115 cells were detected after decontamination. In addition, no specific colonies were seen after a second elution with sterile 0.9% NaCl solution 24 hours after the first sampling round.
Experiments were also carried out with S. cerevisiae-INVSc1 using the same elution scheme as P. pastoris-GS 115. A single second elution of the system 24 hours after decontamination was performed. All samples tested negative for specific CFUs.
Challenging the NGC System with a Combination of E. coli-B13, P. pastoris-GS 115, and S. cerevisiae-INVSc1
Equal volumes were mixed and the individual cell count was as follows: E. coli-B13, 2 x 107 CFU/ml; P. pastoris-GS 115, 4 x 106 CFU/ ml; S. cerevisiae-INVSc1, 4 x 106 CFU/ml. The system was infected and, 24 hours later, samples were taken to confirm CFU data. All three organisms grew extremely well under these conditions and the sheer number of organisms posed an elevated challenge to the sanitization protocol, which was applied unchanged. Sampling with sterile 0.9% NaCl solution was carried out immediately after decontamination and was repeated again 120 hours later. During those 120 hours the NGC System was stored in 0.9% NaCl solution instead of the typical 20% ethanol.
Inspection of agar plates from the first sampling was carried out after 120 hours and two specific events were recorded. There were 10 CFUs (yeast) in 200 ml in the flow path from the buffer blending valve to the waste position of the inject valve (5 CFU/100 ml). An additional 20 CFUs (E. coli-B13) in 450 ml were found in the flow path from the column switching valve to the outlet valve of the system (4.4 CFU/100 ml). In both instances the CFU level was below the GMP requirements (10 CFU/100 ml) for the manufacture of water for injection (WFI). Although the system was in 0.9% NaCl solution for 120 hours, the agar plate inspection of the second sampling was negative and no further specific growth events were observed.
Endotoxin Testing
During the course of the project the NGC System was treated with E. coli-B13 solutions multiple times. Gram negative bacteria such as E. coli-B13 release endotoxins into the surrounding environment upon cell death. Since bacterial endotoxins are potent inflammatory agents, it is critical that they do not accumulate in a chromatographic system. They can be removed with a sanitization agent like NaOH. Endotoxins are very stable lipopolysaccharides and may require cleaning agent incubation as long as 12 hours (Table 2).
Table 2. Endotoxin testing with three panels. Sample panels included A (before contamination), B (after E. coli-B13 contamination of the NGC System and sanitization with NaOH for 1 hour), and C (sanitization with NaOH for 18 hours). Sampling was performed in sections (from left to right). Sampling solution was 0.9% NaCl.
| Microbial Test | Path 1 (5 ml) | Path 2 (5 ml) | Path 3 (10 ml) | Path 4 (10 ml) | Path 7 (10 ml) | |
| A | Endotoxin testing, EU/ml | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
| B | Endotoxin testing, EU/ml | <0.1 | <0.1 | <0.1 | <0.1 | <0.5 |
| C | Endotoxin testing, EU/ml | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
Conclusion
Modern chromatography systems such as Bio-Rad’s NGC System, utilize automated processes that run user-defined cleaning methods for the system. This along with the easy access and high utility inlet values provide users with a convenient and effective flushing of the system so it can be ready for subsequent runs. Establishing a routine maintenance schedule should keep your chromatography system running effectively for years.
This study clearly demonstrates that massive microbial contamination of a comprehensively equipped NGC Chromatography System can be reduced to as low as 1 CFU/10 ml effluent. The cell types used for these challenge experiments represent a common class of organisms utilized for the expression and development of protein-derived therapeutic drugs.
The results demonstrated excellent cleaning efficiency under extreme contamination. Although a few CFUs of the challenging organisms were detected after infection with an E. coli-B13 and yeast cocktail, a minimum of 6 log reductions in CFUs was still achieved and the levels were well below the cGMP mandated 10 CFU/100 ml.
In addition, endotoxin removal was investigated and a contact time for decontamination with NaOH was successful with an incubation time greater than 1 hour. Results such as these should enhance confidence in the NGC System, as pharmaceutical researchers establish and validate cleaning processes.