What do X-ray crystallography, protein binding studies, and antibody production have in common? They all require protein samples of high purity. Although engineering an affinity-tagged version of a protein is a typical first step for easy purification, tags can cause problems for some applications. The task of purifying untagged proteins, daunting to many, is made easier using the Bio‑Rad NGC Chromatography System and prepacked EconoFit Columns. Together, these tools allow researchers to automate purification and significantly accelerate workflow development.
High purity protein samples are essential for many research applications, from protein structure determination and biochemical characterization to antibody production. A common purification approach is the use of affinity tags such as histidine or glutathione-S-transferase tags. These tags increase the throughput and efficiency of the protein purification workflow, but affinity tagging is not always a viable option.
Some proteins are unstable, inactive, or exhibit altered activity once tagged. Cleavable affinity tags can sometimes address these issues, however, this adds extra cost, and/or the protein of interest may require posttranslational modifications that do not permit recombinant expression. Thus, the protein needs to be isolated from a natural source as native protein. When the use of affinity tags is not possible, researchers often settle for lower purity protein rather than investing additional time and labor to extensively explore purification options for untagged proteins.
Exhaustive exploration is often daunting and can be impractical. Identifying an ideal purification workflow requires optimization of multiple variables, including column resin, pH, and elution buffer gradient (%B), at each step (Figure 1). This means that many runs are required at each purification step to identify the purification workflow that yields protein of the desired purity.
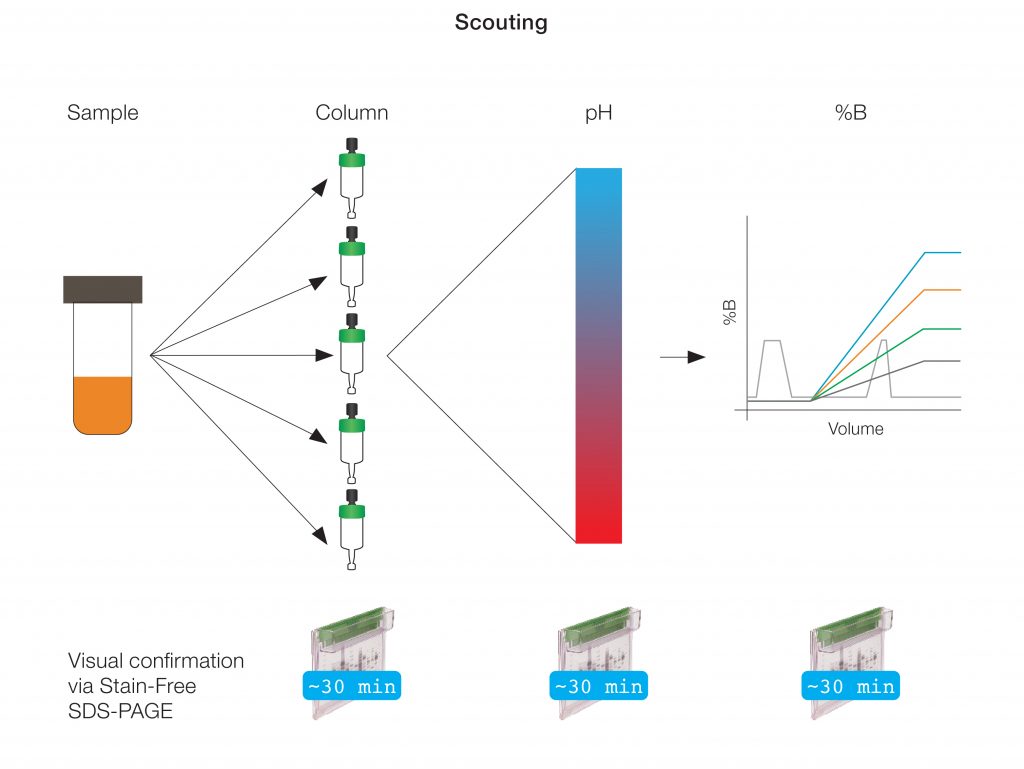
Fig. 1. Protein purification workflow scouting.
Automated Condition Testing for Easy Optimization
Traditionally, the optimization process requires the setting up of individual programmed runs for each varied column, pH, and %B condition. The NGC Chromatography System’s ChromLab Software, however, allows automation and acceleration of this cumbersome process. The scouting and multivariable scouting features in ChromLab Software can be used to easily prepare a series of methods designed to interrogate many experimental conditions in an automated fashion.
The series of methods created will control different components of the NGC Chromatography System, such as the Sample Pump, which directly injects sample onto the column, the Buffer Blending Valve, which allows scouting of pH and %B, and the Column Switching Valve, which can automatically switch between as many as five different columns. All resulting chromatograms can be evaluated in ChromLab Software through peak integration as well as overlaying all traces for a direct run-to-run comparison.
A Practical Test of Rapid Optimization
In our featured study, the NGC Chromatography System allowed us to scout four ion exchange columns without user intervention, with the same set-up and instrument programming time required for just one traditional run. For this test, we purified prancer purple, a chromogenic protein, without affinity tags. Multiple pH conditions were explored without the need to make and titrate individual buffers for each tested pH. Finally, automated %B scouting enabled us to improve the final purity of protein.

The results illustrate that small changes in resin chemistry or pH can have drastic effects on protein binding, and that changes in the elution buffer pH or gradient can greatly impact purity of the eluted protein. The study demonstrates that column scouting is essential, even when a specific resin chemistry, such as anion exchange, has already been chosen.
Resins designed for similar use can show very different binding efficiencies for a given protein. A resin that performs well for one protein may or may not work for another. For instance, even though Nuvia Q and UNOsphere Q Resins are both strong anion exchange resins with trimethylamine functional groups, prancer purple bound only to the Nuvia Q Resin. This is due to differences in resin bead chemistries and size.
With EconoFit Columns, a wide range of Bio-Rad resins are available in prepacked format for scouting applications and to facilitate identification of the optimal resin for a given protein.
Assessing Protein Yield and Purity
A further challenge of purification workflow development is evaluating which methods yields the best results. Identifying purification fractions that contain the highest amount of protein of interest, while containing only few contaminating proteins can be labor intensive. This is especially true when multiple conditions, and therefore, many fractions must be scouted.
Traditionally, fractions are chosen based on A280, tracked throughout the chromatography run, and then analyzed by SDS-PAGE to assess sample purity. In our study, a complementary measure of purity, the purity quotient difference (PQD), is introduced to inform method development when the protein of interest and contaminants absorb light at different wavelengths.
Most proteins contain tryptophan residues and thus absorb light at a wavelength of 280 nm. Chromogenic proteins, fluorescently tagged proteins, or metal-bound proteins such as hemoproteins, as well as contaminants such as DNA, absorb light at also other wavelengths. Using an NGC Chromatography System with a Multi-Wavelength Detector (included in all NGC Plus and Discover Systems), it was possible to distinguish the elution profile of prancer purple (525 nm) from the elution profile of contaminating proteins (280 nm). This formed the basis for PQD analysis, which provided a quick and automated approach to screen and narrow the number of fractions needed for SDS-PAGE analysis to confirm sample purity (Figure 2).
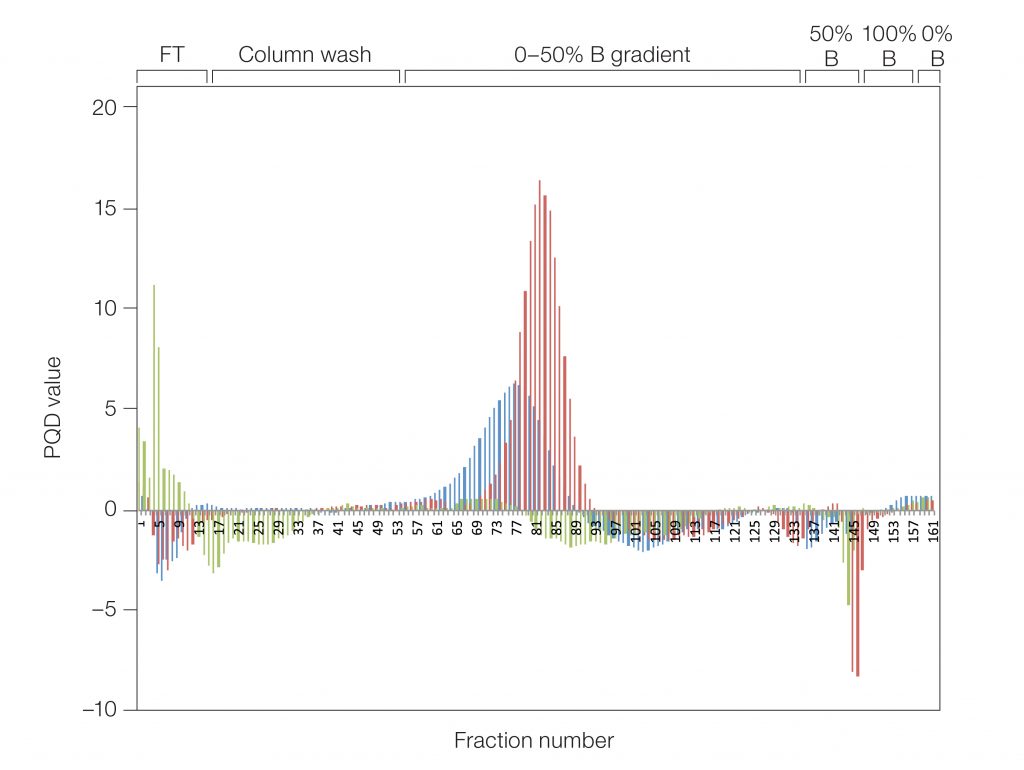
Fig. 2. Histogram showing the purity quotient difference for prancer purple (PQ525–PQ280) per fraction from each column scouting run. UNOsphere Q (■); Nuvia Q (■); Macro-Prep Q (■).
Using Criterion TGX Stain-Free Precast Gels allowed us to resolve and analyze protein samples in less than 30 minutes. This permitted characterization of prancer purple elution patterns from a large number of fractions in a very short time.
As this prancer purple case study illustrates, NGC Chromatography System–mediated automation of these scouting processes enables exhaustive exploration of purification options. Combined with ChromLab Software’s intuitive programming features and preloaded purification templates, the NGC Chromatography System allows the streamlined purification of untagged proteins without compromising protein purity or integrity.
Click here to view the full results from our case study.

