Over the past several decades, our understanding of disease has increased significantly. Since the advent of next-generation sequencing (NGS) in particular, we have been able to unravel the complexity of several diseases and disease states. For example, with these data, we can gain a deeper appreciation for the complexity of disease, down to the expression levels of proteins. However, much of the data generated from a single NGS run lack any known functional role and are often deemed useless. Artificial intelligence (AI), which relies on building a model and applying it to solve a specific problem, has been used in a variety of use cases, including genomics. There are many areas of genomics that can be accelerated using AI and machine learning including sequence analyses, drug development, clinical trial design, process development, and manufacturing.
The Hierarchy of AI
AI, machine learning, and deep learning are terms thrown around with their meanings often conflated. The broadest term is AI, which loosely involves incorporating human intelligence into machines. We come across many uses of AI in our everyday lives, including for example, elevator door sensors. A subset of AI is machine learning, which uses machines to help identify patterns from data rather than follow strict programming rules. Within genomics, this could include using data sets to generate computer models that can make predictions about an individual’s odds of developing a disease or responding to interventions. An even more specialized subset of machine learning is deep learning. The key distinction here is that machine learning algorithms almost always require structured data, while deep learning networks rely on layers of artificial neural networks. These neural networks roughly mimic how our neurons work, with connections (dendrites or axons) between neurons and the transfer of signal. Finally, deep learning refers to the depth of layers between inputs and outputs. Often, there are complex relationships in data that can only be modeled by using multiple (hidden) layers. A neural network can learn relationships between the features that other algorithms cannot easily discover.
AI in NGS
There are three major areas where AI can be used within genomics, spanning across the next generation sequencing workflow — primary, secondary, and tertiary analyses. Before NGS data are generated, researchers decide the theoretical depth of coverage, which is the number of roughly 250 base pair reads needed to obtain optimal coverage across the genome or region (that is, gene of interest). Imagine a pile of tens of millions of reads in order to obtain 20x fold-coverage (the number of times a base has been sequenced) across the genome. AI can be leveraged to determine the optimal sequencing depth based on the desired outcome or result. This takes into account factors like the frequency of variant and similarity of reads at that particular part of the genome. Minimizing the depth of coverage can be beneficial in eliminating extraneous reads, resulting in faster turnaround times and cost savings.
Another area where AI can be applied is mapping or assembly. Assembly refers to a part of secondary analysis where the individual reads (approximately 250 base pairs) are put together to make a de novo genome. The alternative to assembly, is mapping, where reads are assigned to a known and existing genome. Both assembly and mapping are compute intensive. AI models can be developed based on the patterns of existing genomes to both map and assemble more efficiently. Using AI can significantly reduce the time and compute resources required for secondary analysis.
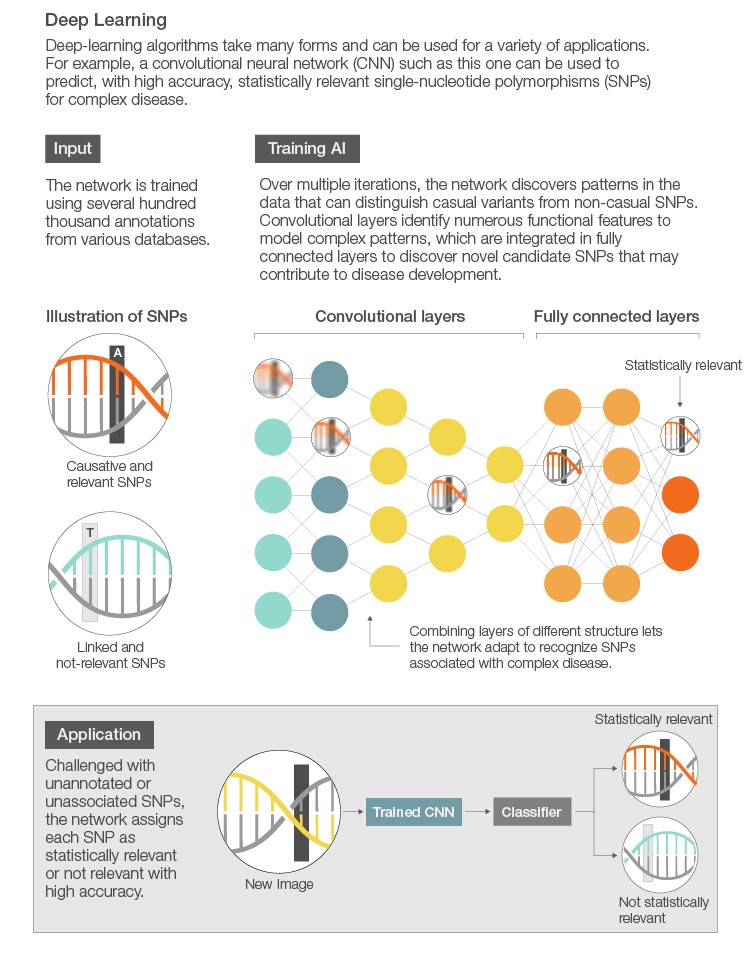
Following assembly or mapping is tertiary analysis, where differences between a known and new genome are interrogated. These differences can be large insertions, deletions, or rearrangements or extremely small single nucleotide polymorphisms (SNPs). Detecting these variations (variant calling) is relatively straightforward and well supported by the open-source community. For example, there are several analysis pipelines, including the Genome Analysis Toolkit (GATK), which make it easy to identify SNPs and small insertions and deletions (indels) in germline DNA and RNA-Seq data. Once variants are called, the next step is to determine which variants are statistically or clinically relevant. Here, AI can be used to detect patterns of variants (mutations) within healthy or diseased patients, for instance, to determine which variants are relevant. Additionally, finding clinically relevant variants is a labor-intensive process. Harnessing the power of AI can drastically reduce the time and effort in finding key clinical drivers.
Clinical Trial Design
One of the most common types of clinical trial designs is umbrella trials, where patients are assigned to a particular treatment arm of the trial based on, say, their type of cancer. For example, patients with lung cancer would potentially be binned together. Alternatively, basket trials test the effect of one drug on a single mutation in a variety of tumor types at the same time. For example, all patients with a specific mutation within BRCA1 would be grouped together, regardless of if they had lung or breast cancer. These studies are extremely promising, as they have the potential to greatly increase the number of patients who are eligible to receive certain drugs relative to other trials designs. AI has been used to help determine the specific molecular makeup (biomarker) for a particular drug or treatment, potentially increasing the number of patients who can receive that treatment.
The Promise of AI
The extremely cumbersome drug development process, combined with a lengthy and often difficult drug approval process, forces researchers to be innovative to overcome these obstacles. Across analysis pipelines, AI tools can be leveraged to accelerate research and development, process development, manufacturing, and clinical trials. As we uncover the true complexity of diseases, we realize determining patterns of biomarkers is extremely difficult. The power of AI is that we may not need to fully understand disease pathways or mechanisms in order to effectively treat them. As in the case of deep learning, the hidden layers don’t need to be described. In fact, we don’t even need to completely understand which inputs (variables) are deterministic.
AI tools will only continue to improve over time, as will the number and quality of applications specifically within drug development, resulting in greater efficiencies for BioPharma companies in terms of time to market and cost. Process development and manufacturing, among other areas within BioPharma, are also leveraging AI. This too will increase over time and extend across additional future applications.


