In 1990, researcher Mary-Claire King and her colleagues linked a region of human DNA with increased risk for breast cancer. Building on this work, researchers soon cloned two breast cancer susceptibility genes, BRCA1 and BRCA2. Certain harmful mutations in these genes correspond with a fivefold increase in a woman’s risk for breast cancer, from about 12% without the mutations to 60% with them, as well as increased risk for other cancers such as ovarian cancer (National Cancer Institute publication, 2009). Today, testing for BRCA1/BRCA2 mutations is a well-established part of genetic screening. Along with constituting an early advance in personalized medicine, the BRCA findings have given cancer researchers an important key: the potential first link in the cellular chain of events leading to a specific cancer.
However, a generation after King made the connection, we are still trying to identify the steps by which mutations in BRCA genes may result in breast or ovarian cancer. Researchers are now trying to describe the mechanisms involved by identifying specific proteins and characterizing their interactions. Spelling out exactly which proteins act and how they do so promises to translate into potent new therapeutic targets. Ryan Jensen, a researcher in Yale University’s Department of Therapeutic Radiology, is among those mapping the trail that begins with the BRCA genes and may end with a new understanding of cancer and how to treat it.

Assistant Professor of Therapeutic Radiology
Yale School of Medicine
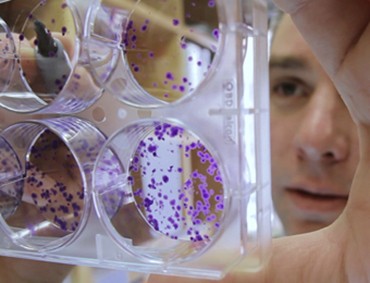
Researcher Ryan Jensen checks the confluency of HeLa cell colonies. Using an HIV lentivirus vector, he will transfect these cells to express BRCA2 protein to use in a series of biochemical assays. By characterizing the BRCA2 protein, Jensen hopes to illuminate its role in DNA repair mechanisms and discover therapeutic targets for when these mechanisms go wrong.
A link to DNA damage and repair
Of the two breast cancer genes, Jensen focuses on BRCA2. Among the functions encoded by this gene is a major role in DNA repair, especially in homologous recombination, the cell’s response to the most catastrophic type of DNA damage, double strand breaks.
“We like to think of our DNA as being static, safe within the nucleus of a cell,” Jensen says. “But it’s actually experiencing constant damage. As the cell divides it has to deal with this each time it replicates the DNA. Every once in a while the DNA gets broken, actually physically separated, through a double strand break.”
Protein by protein, Jensen wants to lay out the response to these breaks and the part played by BRCA2. Understanding how this response works and what goes wrong at the molecular level could illuminate how errors sneak into cellular DNA, creating the hallmark genomic instability observed in cancer cells, as well as showing why these diseased cells are often so hard to stop once they start proliferating.
“Do we need mutations in DNA repair genes to drive cells from being a normal cell to being a cancer cell?” Jensen asks. “Then, once you have the cancer cell and you start treating those tumors, is it the DNA repair genes that start getting up-regulated and repairing all that damage while we’re trying to kill the tumor cell?”
The problems of a large and fragile protein
For those trying to answer these questions, BRCA2 posed a major challenge: its size. It’s about three times larger than the average gene, and the BRCA2 protein it encodes is also quite large (3,418 amino acids, or about 390 kilodaltons). In addition, in vivo the protein generally exists in complexes; on its own, without these stabilizers, it’s fragile and unstable. Because of these hurdles, researchers could work only with truncated versions of the BRCA2 protein, which greatly limited their ability to model its function.
As a postdoc in Stephen Kowalczykowski’s lab at the University of California, Davis, Jensen spent four years in the cold room trying different purification strategies for BRCA2. Ultimately, he used a maltose-binding affinity tag to stabilize the protein and human epithelial kidney cells to express it. He and his colleagues published their results in a 2010 Nature article (Jensen et al. 2010). “In retrospect, it seems trivial that adding the tags to the protein and expressing it in human cells would be a feasible strategy,” says Jensen. “But it was really the difference between failure and success.”
Characterizing BRCA2’s behavior in double strand break repair
Purifying BRCA2 was a big breakthrough in that it opened the door to characterizing the full protein’s biochemistry. Jensen’s 2010 paper not only described how he and his colleagues purified BRCA2, but also pulled back the curtain further on how BRCA2 participates in homologous recombination, especially how it interacts with a DNA repair protein called RAD51 in bringing the damaged DNA’s sister chromatid to the rescue.
“RAD51 is one of the central players in homologous recombination,” Jensen explains. “After a double strand break, each of the DNA ends hanging free gets resected to reveal a three-prime single-stranded tail. RAD51 forms a filament that wraps around the single-strand DNA, extending and stretching it. The RAD51-nucleoprotein filament then invades a duplex donor DNA and searches for homology, looking for a base pair that it can be put into register with. That initiates the process of homologous recombination. What we quickly realized from our biochemical assays was that BRCA2 could actually take RAD51 like a cargo, binding multiple RAD51 proteins and depositing them to a DNA double strand break.”
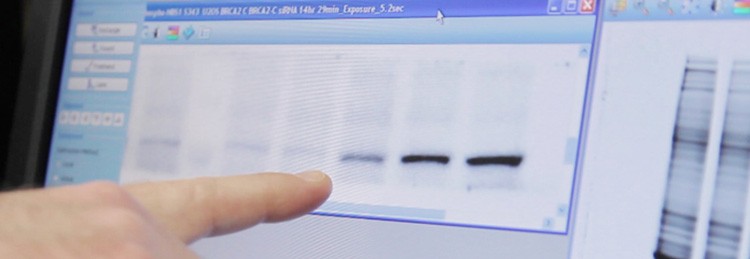
Using the ChemiDoc MP imaging system, Jensen points to bands showing BRCA2 expression levels in relation to siRNA-mediated knockdown of RAD51 paralog proteins. Imaging total proteins on his gel (at right) allows him to normalize his western blot quantitation to a more accurate reference than housekeeping proteins.
The search for more proteins interacting with BRCA2
Following his work characterizing BRCA2 and RAD51 interactions, Jensen has turned his attention to a group of five RAD51 paralog proteins, which were identified in the late 1990s. “These proteins have similar sequence homology, similar amino acids, but they’re different than RAD51,” says Jensen. Looking first to interactions at the genetic level, Jensen utilized RNA interference to knock down expression levels for each of these paralog proteins. In doing so, he found evidence that the paralogs work in the same pathway as BRCA2 (Jensen et al. 2013).
Having established their genetic epistasis with BRCA2, Jensen now wants to turn to the direct interactions of the paralogs and BRCA2 to better understand their biochemistry. “Biochemistry really allows us to see what these paralog proteins are doing,” he says. “Do they collaborate with BRCA2? Do they enhance this RAD51 filament formation? Are they a part of the RAD51 filament? At this point it’s a completely open question. The only way to answer it is to purify these proteins and put them together in a test tube. Then, we can use these in vitro assays to ask questions such as, is there a specific order that we need to put these proteins in to see the exact mechanisms of action?”

Jensen and student Tiffany Nguyen examine images with the ChemiDoc MP imaging system. The V3 Western Workflow gives them multiple quality-control checkpoints so they can verify protein separation and transfer, then quantitate western blots using Image Lab software.

Jensen credits the V3 Western Workflow for revealing a pattern of unexpected bands in gels showing BRCA2-RAD51 paralog protein interactions. He intends to analyze these unknown bands using mass spectrometry to see if they indicate a novel interaction.
A better way to measure proteins
Running these assays entails making many different protein combinations and many different measurements to carefully test interaction effects and slowly model the mechanisms. The goal is to map the sites of interaction for BRCA2 and a given paralog. To do this, Jensen starts with chunks of BRCA2 protein, then gradually looks at smaller and smaller sections, narrowing in on the exact site for a given interaction. This means running gel after gel. Jensen reports that, in a busy week, his lab might run 30, 40, or even 50 gels. In both his current work with RAD51 paralog biochemistry, and in the genetic interaction studies underlying it, Jensen has had the help of an innovative new system for measuring protein levels: Bio-Rad’s novel V3 Western Workflow™.
Combining three innovative systems, TGX Stain-Free™ precast gels, the Trans-Blot® Turbo™ protein transfer system, and the ChemiDoc™ MP imaging system, this new western blotting workflow offers Jensen a better way to analyze his protein experiments, particularly when it comes to quantitation.
“Using the stain-free gels in conjunction with the ChemiDoc system allows us to more accurately quantitate our western blots,” says Jensen. This improvement results from the
V3 Western Workflow’s use of total protein normalization as a loading control, a significant change from the traditional method.
“Most people normalize their western blots with housekeeping proteins (HKPs) — tubulin, actin, GAPDH — these are probably the most common,” Jensen explains. “The downside of doing that, and you see this over and over in the literature, is that they usually blow out the signals from the actin or tubulin so that it’s no longer in the linear range of detection of the instrument. So you see these huge fat bands of actin and tubulin all along the bottom of the western blot image. If you’re trying to compare your protein of interest and you’re trying to normalize to this blown out, saturated signal of actin or tubulin, you’re never going to get a quantitative result.”
Rather than the problematic HKPs, the V3 Western Workflow pairs stain-free technology with the ChemiDoc MP imaging system to detect the total proteins in a given gel. The technology works by seeding the gel with UV-excitable molecules that fluoresce when bound to tryptophan residues. The sensitivity is comparable to a Coomassie stain, but because it works through imaging rather than staining, total protein can be detected on the same gel used for the western blot. Thus, the measure of total protein can now function as a reference for western blot quantitation.
As Jensen describes, “[Using the Image Lab™ software], you can box each lane and get a representation of all the proteins in the lane. That’s a much more dynamic range than just looking at actin or tubulin. I think scientifically that’s a better way to go about it, a better way to normalize your protein of interest if you really want to be precise and compare protein expression from different types of DNA damage treatments. You really want to normalize to something that’s going to be representative of what’s going on with all these other proteins in the cell.”
Using this total range of protein as a reference for quantitation also alleviates uncertainties stemming from the specific cellular phenomena Jensen’s investigating. “In our case, we’re treating cells with DNA damage,” he notes. “If actin or tubulin are in any way affected by DNA damage, that could interfere with the analysis of our results.”

Stain-free gel technology works using markers in the gel that bind to tryptophan molecules and react to fluorescent imaging, enabling all proteins on the gel to be visualized without a Coomassie or similar stain.
An improved workflow also opens the door to discovery
Along with the more accurate and reliable quantitation, Jensen also appreciates the efficiencies the V3 Western Workflow brings to measuring proteins. The stain-free gels run faster, in as little as 15 minutes, while imaging takes only 3 minutes. The ability to image total proteins also eliminates having to strip the blot and reprobe it to establish the HKP reference, a time-consuming and error-prone step. Instead, using the ChemiDoc MP imaging system introduces greater quality control at more points in the process.
“You can image your gel right after you run it and see all the proteins in your lane,” Jensen says. “Then you can transfer that to your nitrocellulose membrane or your PVDF membrane, and then again you can image that membrane to see that your transfer worked; that’s another level of quality control.”
These quality control steps may also provide new opportunities for discovery, since the researcher now has more points to make observations. Jensen illustrates one instance: “When we started visualizing [our] gels on the ChemiDoc system I noticed some other bands showing up besides BRCA2 and the paralog proteins that I was anticipating. I noticed that the pattern was different depending on which piece of BRCA2 we were isolating. So perhaps we’re identifying other proteins that interact with specific regions of BRCA2. That’s a really cool, unanticipated side effect of using the stain-free gels.”
Going from a band on a gel to the patient in the clinic
Along with illuminating the biochemical processes of DNA repair, Ryan Jensen’s work with BRCA2, RAD51, the RAD51 paralogs, and the other proteins he may discover along the way has significant clinical potential. “It seems like a huge disconnect to go from looking at some protein band on a gel to the patient in a clinic, but it’s not that far-fetched,” Jensen emphasizes.
Having a better way to do western blotting makes a difference in this effort. “If we can see that new protein, if that’s a potential new therapeutic target, that’s what we really want,” Jensen continues. “This is one way in which this system excels. We want to identify that protein as quickly as possible and we want to make sure we’re doing it right and we’re doing it rigorously and we’re doing it quantitatively.”
References
National Cancer Institute (2009). BRCA1 and BRCA2: cancer risk and genetic testing. Cancer.gov. Accessed May 13, 2013.
Jensen R et al. (2010). Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678-683.
Jensen R et al. (2013). BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair 12, 306-311.

