Preamplification is a powerful technique that enables qPCR analysis for many targets from limited samples. However, it is frequently treated as a necessary evil as many fear that it may introduce bias into their experiments, leading to the generation of inaccurate results. Preamplification is also sometimes misunderstood as being a magic bullet to boost qPCR sensitivity. This article addresses these issues as well as others, so that you can be confident when using preamplification in your qPCR experiments.
1. What is preamplification and how is it useful for qPCR?
PCR-based preamplification is a method used to increase the concentration of a specific panel of targets in a sample prior to qPCR analysis, reducing the required sample input for multi-target qPCR experiments. Preamplification is essentially a highly multiplexed PCR reaction performed for a limited number of cycles using the same primer sets that will be used in the downstream qPCR reaction. By using a reagent designed for preamplification with a limited number of PCR cycles, optimal amplification efficiency can be maintained for each target, which is essential to preventing the introduction of bias into the qPCR analysis. With just 10–14 cycles of preamplification, the concentration of each target is boosted by 1000-fold or more, providing enough preamplified sample to analyze all of the targets by qPCR without compromising the sensitivity of the qPCR analysis. As a rule of thumb, preamplification is useful any time the amount of sample available limits the number of targets that can effectively be analyzed.
2. Will preamplification introduce bias to my experiment?
The introduction of bias into qPCR experiments is a primary concern when considering the use of preamplification. Okino et al. (2015) investigated and discussed three types of preamplification-associated bias: amplification, fold change, and dynamic range.
Amplification bias occurs when PCR efficiency is suboptimal for some targets, leading to the apparent under- or overrepresentation of those targets in the preamplified sample. To minimize the introduction of amplification bias, a preamplification reagent must be able to maintain optimal PCR efficiency across all of the targets over the course of the preamplification. Reagents designed for preamplification typically have bias specifications based on amplification bias.
More importantly, to truly obtain accurate results with preamplification, you also have to consider fold-change bias, where the fold difference in target abundance measured between two samples using preamplification deviates from the actual fold difference. The presence of amplification bias may not necessarily mean that fold-change bias has also been introduced. Okino et al. investigated fold-change bias using the ERCC set of exogenous RNA spike-in controls (for more information about ERCC controls, see nist.gov/programs-projects/external-rna-controls-consortium). Using these controls, they were able to analyze a pair of samples where RNAs were present at differing known levels, allowing the fold difference between the samples to be measured and compared with and without the use of preamplification. They tested preamplification with both 88 and 355 targets and found no systematic differences in the fold difference measurements related to the use of preamplification (Figure 1).
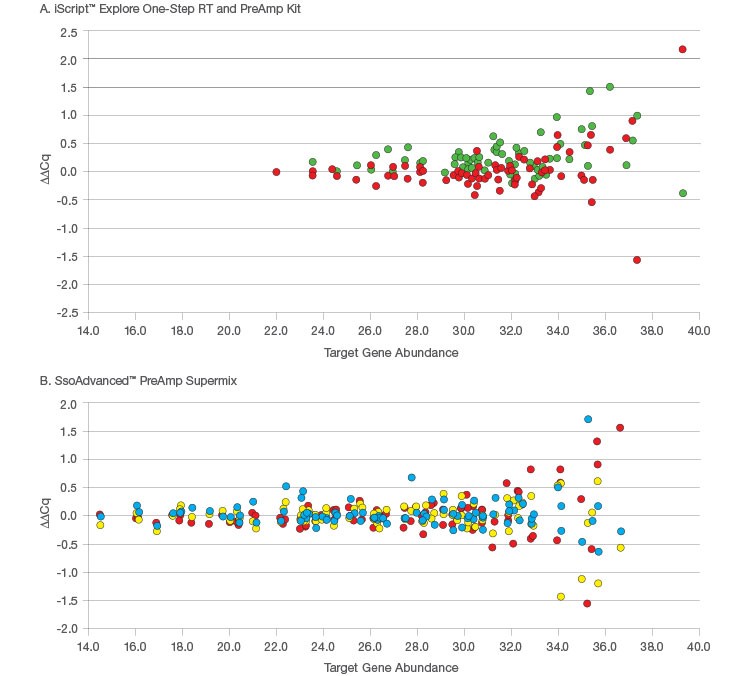
Fig. 1. Analysis of fold-change bias introduced by preamplification. The ΔΔCq results show the difference between the measured and expected fold-differences between samples measured by RT-qPCR with and without preamplification. A, data for iScript Explore One-Step RT and PreAmp Kit where 81 targets from the PrimePCR Pan-Cancer lncRNA qPCR Array were analyzed in 10 ng and 40 ng of Universal Reference RNA (Agilent) with preamplification (![]() ) and without preamplification (
) and without preamplification (![]() ). B, data for SsoAdvanced PreAmp Supermix from Okino et al. where 88 ERCC ExFold RNA Spike-In Mixes (Life Technologies) were assessed using 88-plex preamplification (
). B, data for SsoAdvanced PreAmp Supermix from Okino et al. where 88 ERCC ExFold RNA Spike-In Mixes (Life Technologies) were assessed using 88-plex preamplification (![]() ), 355-plex preamplification (
), 355-plex preamplification (![]() ), and no preamplification (
), and no preamplification (![]() ).
).
Okino et al. also investigated dynamic range bias to determine whether target concentration has a systematic effect on the accuracy of qPCR results generated using preamplification. They found that when they performed preamplification for 20 cycles, some highly abundant targets produced qPCR results with extremely low Cq values, around 5. Because it is difficult to properly baseline the fluorescence data for targets that amplify this early, these qPCR data cannot be reliably used. When preamplification is performed for a more moderate 10–14 cycles, it is far less likely that dynamic range bias will be encountered.
Because preventing both amplification and fold-change bias during preamplification is inherently dependent on maintaining optimal PCR efficiency for all targets during the course of preamplification, the introduction of bias can be avoided or minimized by using a preamplification reagent designed for this purpose. Bio-Rad’s preamplification kits utilize the patented* Sso7d fusion polymerase to this end. The enzyme contains an Sso7d double-stranded DNA binding domain that confers high processivity to the polymerase, allowing it to achieve efficient preamplification of many targets simultaneously (specifically, 100 targets with the iScript Explore One-Step RT and PreAmp Kit and 400 targets with the SsoAdvanced PreAmp Supermix).
3. How do I calculate the level of target enrichment achieved through the use of preamplification?
The expected level of target enrichment achieved via preamplification can easily be calculated based on the volume of sample input, the number of PCR cycles used for preamplification, and the final volume of the diluted preamplified sample. For example, when analyzing 10 µl of RNA using the iScript Explore Kit with the recommended protocol, you can expect to achieve an approximate 328-fold enrichment of each preamplified target (assuming 100% PCR efficiency):
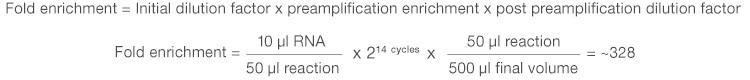
4. Does the use of preamplification improve qPCR sensitivity?
While the use of preamplification will produce significantly earlier Cq values in qPCR due the level of target enrichment, it is not a magic bullet to boost qPCR sensitivity (that is, the minimum number of target copies that can be detected in the qPCR reaction). The limit of detection for qPCR is already close to single copy in a well-optimized qPCR assay, though stochastic effects make it difficult to accurately deliver and/or quantify less than about 10 copies per reaction. When preamplification is used, the limit of detection is instead based on the lower limit of target that must be added to the preamplification reaction such that it can still be detected by the downstream qPCR reaction. Given that both qPCR and preamplification use PCR for amplification, the limit of detection should be similar with or without the use of preamplification.
Even though preamplification in itself does not improve qPCR sensitivity, the use of preamplification generally allows the modest increase of the effective amount of sample analyzed by qPCR, with the entire amount of sample input used in the preamplification reaction in turn being interrogated by each downstream qPCR reaction. For example, up to 10.5 µl of RNA can be used as input for a one-step preamplification reaction with the iScript Explore Kit, thus allowing the effective analysis of 10.5 µl of RNA by each downstream qPCR reaction. In contrast, if you were to analyze the same RNA sample using a standard RT-qPCR workflow, the effective amount of RNA sample analyzed in each qPCR would only be 0.7–1.5 µl given typical reaction volumes (Figure 2). So while preamplification in itself doesn’t increase the sensitivity of qPCR, it does maximize the amount of sample effectively analyzed by qPCR while simultaneously allowing an increase in the number of targets included in the analysis.
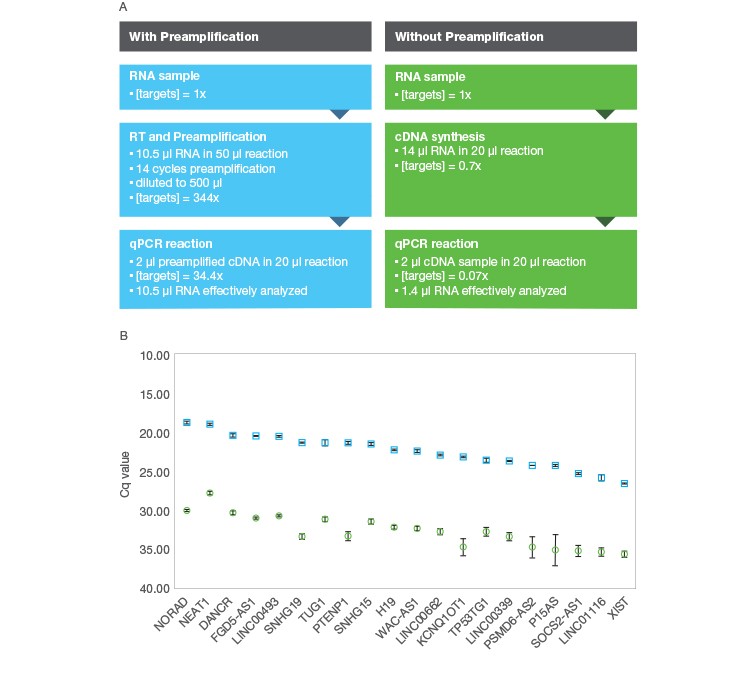
Fig. 2. Comparison of RT-qPCR workflows attempting to maximize sensitivity with and without preamplification. A, shows a comparison of theoretical target levels throughout the course of RT-qPCR using the iScript Explore Kit (■) or without preamplification using the iScript gDNA Clear cDNA Synthesis Kit (■). While the cDNA synthesis reaction can accommodate 14 μl of RNA input, there is no enrichment during the cDNA synthesis, so the qPCR effectively analyzes just 1.4 μl of RNA. In contrast, the preamplification workflow effectively analyses the full 10.5 μl of RNA added to the iScript Explore reaction leading to the effective analysis of 7.5-fold more sample with preamplification. B, results for 20 lncRNA analyzed with both workflows. 1 ng of Universal Reference RNA (Agilent) was used as input for the iScript Explore Kit while 1.33 ng was used with the iScript gDNA Clear cDNA Synthesis Kit. The average Cq across targets for the iScript Explore Kit was 22.6 while the average for the iScript gDNA Clear Kit was 32.7 to analyze from the compared RT-qPCR workflows. The average ΔCq between the workflows was observed to be 10.1. (Note: the theoretical 491-fold difference in target concentration between the methods at the qPCR level is expected to produce a ΔCq of 8.9, but the two workflows use different RT priming methods that are not taken into account.)
5. How do I validate the use of preamplification for my set of targets?
The introduction of amplification bias through the use of preamplification is easy to assess by comparing qPCR results with and without preamplification. You would expect to see a shift (ΔCq) in qPCR results that is consistent with the level of fold enrichment from the preamplification, assuming the same amount of sample input is used for both methods (that is, if preamplification is performed with 2 µl of cDNA, then the comparison should be made to qPCR with 2 µl of cDNA used as input). Generally, the results are acceptable when the ΔCq is within ± 0.75 of the expected value.

While measuring amplification bias is less relevant to the accuracy of the qPCR results than testing for fold-change bias directly, it is easier to accomplish because it does not require testing a pair of samples with a known fold-difference for the targets of interest. If you want to also test for fold-change bias, one way to do so would be to measure the fold change detected for a given sample dilution, as was done with the iScript Explore Kit in Figure 1.
In addition to testing for bias, it is also good practice to include a negative control and melt curve analysis if using SYBR® Green. The negative control will allow identification of any potential nonspecific amplification products that may occur in the absence of template and help ensure that you select a suitable upper Cq limit for determining positive results (for example, the iScript Explore Kit recommends a Cq limit of 29–33 depending on the protocol used). Melt curves obtained using preamplification should match those obtained without its use.
6. Can I confirm that preamplification is working correctly?
With the launch of the iScript Explore Kit, Bio-Rad also introduced the PrimePCR™ PreAmp Control Assay, which allows you to measure the preamplification efficacy for each sample. This control assay uses a synthetic template and primer set to perform preamplification on a segment of the synthetic template. The assay then uses two qPCR primer pairs to compare the levels of the preamplified and non-preamplified segments, which results in a ∆Cq equal to the number of cycles used for preamplification (Figure 3). This control assay is ideal for confirming that preamplification has worked effectively for individual samples.
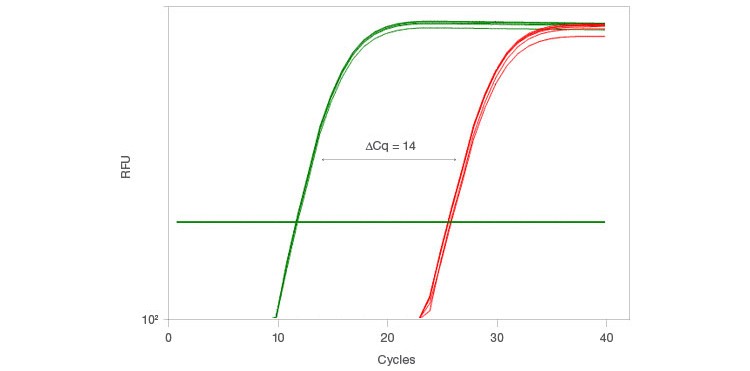
Fig. 3. The PrimePCR PreAmp Control Assay monitors preamplification performance when using the iScript Explore One-Step RT and PreAmp Kit. The control assay uses two primer pairs, PAQ1 (—) and PAQ2 (—), to compare results from a synthetic template where PAQ1 is preamplified while PAQ2 is not preamplified. The ΔCq obtained by comparing the results of PAQ1 and PAQ2 should be equal to the number of cycles used for preamplification. Here, preamplification was performed for 14 cycles using the iScript Explore Kit.
7. What considerations are needed when interpreting data from preamplified samples?
When preamplification is used, Cq values in qPCR will shift earlier due to the enrichment achieved during preamplification. In the example given earlier, 14 cycles of preamplification were used to achieve an approximate 328-fold enrichment from 10.5 µl of an RNA sample. If 2 µl of the preamplified sample were analyzed by qPCR, then the expectation is that ~66 copies of each target is added to the qPCR reaction for every copy of each target that was added to the preamplification reaction. This in turn means that the qPCR results would be shifted ~6 cycles earlier due to the use of preamplification. This value can be used to adjust the upper Cq limit used for determining positive results. If you use a Cq limit of 35 for positive results without preamplification, then the Cq limit would be adjusted to 29 for this example.
8. What should I consider when selecting a product for preamplification?
It is critical to select a preamplification reagent that ensures that all targets in the reaction can be preamplified with optimal PCR efficiency. We recommend the use of either SsoAdvanced PreAmp Supermix or the iScript Explore One-Step RT and PreAmp Kit.
SsoAdvanced PreAmp Supermix allows for preamplification of up to 400 targets from DNA or cDNA samples,making it ideal for samples with limited template DNA — start with as little as 100 pg of cDNA or genomic DNA from any precious or limited sample such as stem cells, laser capture microdissections, and formalin-fixed paraffin-embedded tissues (FFPE).
The iScript Explore One-Step RT and PreAmp Kit enables the generation of preamplified cDNA for up to 100 targets directly from RNA by combining reverse transcription and preamplification into a single step. The kit also integrates an effective genomic DNA removal step, helping ensure only the transcripts of interest are detected by qPCR.
Both products utilize the patented Sso7d fusion polymerase to perform preamplification in an unbiased manner. The unique structure of this Sso7d fusion polymerase provides several significant advantages over ordinary DNA polymerase, including increased processivity, effective amplification in GC-rich regions or strong secondary structure, efficient amplification of longer products, and superior tolerance to PCR inhibitors.
PrimePCR PreAmp Assays are available for the preamplification of over 28,000 gene-specific targets, including common human genes and targets in 12 other widely studied organisms. PrimePCR assays were designed by prioritizing the gene regions most commonly found in transcript variants, using strict design criteria to ensure optimal qPCR results for each target.
Visit Bio-Rad.com for more information about Bio-Rad’s PCR and qPCR Reagents.
References
Okino ST et al. (2015). Evaluation of bias associated with high-multiplex, target-specific pre-amplification. Biomol Detect Quantif 6, 13–21.
Additional Reading
Kibschull M et al. (2016). Quantitative large scale gene expression profiling from human stem cell culture micro samples using multiplex pre-amplification. Syst Biol Reprod Med 62, 84–91. PMID: 26237078
Quinlan A (2015). Too Many Questions, Too Little Sample: Developing a Real-Time PCR Workflow for Monitoring Gene Expression in Limited Samples. Bioradiations, June 9, 2015.
Yadav G (2015). So, How Can the Sso7d Fusion Polymerase Technology Help Your PCR? Bioradiations, December 8, 2015.
Zeka F et al. (2016). Straightforward and sensitive RT-qPCR based gene expression analysis of FFPE samples. Sci Rep 6, 21418. PMCID: PMC4761903
Zou J et al. (2017). Rapid detection of donor cell free DNA in lung transplant recipients with rejections using donor-recipient HLA mismatch. Hum Immunol 78, 342–349. PMCID: PMC5595356
SYBR is a trademark of Life Technologies Corporation.
* U.S. patents 6,627,424; 7,541,170; 7,560,260.

