All of us could use some help for something or other. The DNA polymerases used for PCR are no exception. Some need help with nonspecific amplification at certain temperatures, some with specificity and fidelity, and some with processivity. Several approaches have been taken to improve each of these aspects. Notable among these is the fusion of the double-stranded DNA (dsDNA) binding protein Sso7d to DNA polymerases. Here we discuss how fusion to Sso7d helps DNA polymerases increase their processivity exponentially, and how that in turn makes our PCR and qPCR reactions faster and more efficient.
DNA Polymerases
Nature has endowed cells with DNA polymerases to execute the transmission of genetic information during the cell cycle, and consequently aid in the preservation and sustenance of life. Several families of DNA polymerases have been identified based on their structure as well as amino acid sequences (reviewed in Laos et al. 2014). Identified earlier and most common among them are DNA polymerase families A, B, and C, corresponding to DNA polymerase I, II, and III from E. coli (Ito and Braithwaite 1991, Braithwaite and Ito 1993). Other families such as D (involved in polymerase activity and proofreading in Archaea), X and Y (involved in repair), RT (reverse transcriptase involved in reverse transcription) also exist and play replicative and nonreplicative roles. The most studied naturally occurring DNA polymerases belong to family A, which includes enzymes such as E. coli Pol I, T7 polymerases, and Taq polymerase; and family B, which includes T4 DNA polymerase, RB69 DNA polymerase, and Archael polymerases such as Pfu and DeepVent. The thermally stable DNA polymerases from both families are exploited widely in PCR-based applications. Among the naturally occurring polymerases, some are replicative (for example T7 or DNA Pol III, involved in DNA synthesis) and some are nonreplicative (for example Taq, involved in replication or damage repair). The replicative polymerases are essential for DNA replication and are the primary focus of our discussion here.
DNA polymerases, which are the integral components of the intricately woven replisomes responsible for replication, bind to template DNA and execute extension of a new DNA strand by incorporating thousands of nucleotides with great fidelity (Figure 1). The ability of these enzymes to incorporate nucleotides and extend the strand depends on various aspects, but chiefly on their processivity.
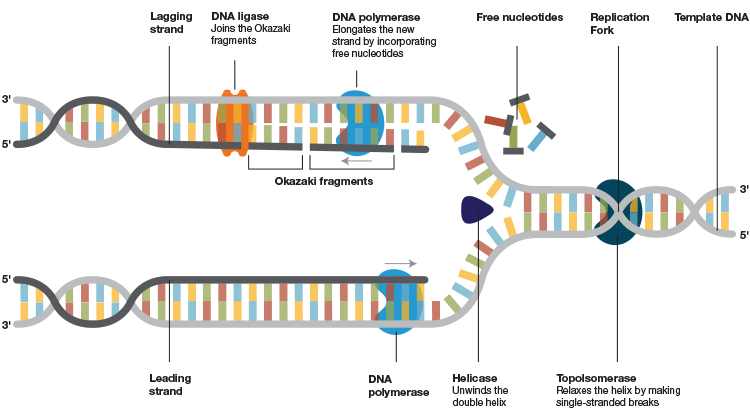
Fig.1. DNA replication machinery. After the helicase unwinds the double helix and the primase assembles the foundation, DNA polymerase binds to the template strand and attaches new complementary nucleotides to the exposed bases on the opened-up template to form and extend the new strand. It moves along the DNA template and keeps incorporating the free nucleotides until it reaches the end of the strand. A stable association of the polymerase to the template DNA is crucial for an effective extension of the new strands. This association is jeopardized in a PCR/qPCR reaction by the nature of the target DNA to be amplified and under challenging PCR conditions.
Processivity of DNA Polymerases — What Does It Mean and Why Does it Matter?
Processivity is the ability of the polymerase to accomplish DNA replication without dissociating from the template and it can be measured by the average number of nucleotides incorporated per binding event. A polymerase with high processivity is desirable and beneficial for achieving high PCR efficiency, especially under challenging PCR conditions. The efficiency of a PCR reaction is defined as the fraction of target molecules copied in one PCR cycle (Alvarez et al. 2007, Lalam 2006). In a properly designed PCR assay under nonchallenging conditions (such as without the interference of PCR inhibitors), target DNA amplification is expected to be achieved with a 99% efficiency (Svec et al. 2015). Most naturally occurring replicative DNA polymerases possess low processivity and are unable to carry out in vitro DNA replication efficiently by themselves. This inadequacy in vivo is offset by associations with various accessory proteins (called processivity factors). Processivity factors assist polymerases in various ways — some by forming complexes with the polymerase itself (for example, thioredoxin), and some by encircling duplex DNA (for example, clamp protein) — thereby ensuring a strong, stable binding with the template (Zhuang and Ai 2010). A stable association of the polymerase with the template DNA is crucial for the unfettered incorporation of nucleotides, and therefore, for an efficient PCR reaction.
Fusion with Sso7d Protein — An Effective Way to Stabilize the DNA and Polymerase Complex during PCR Extension
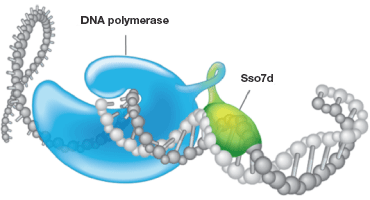
Fig.2. Stabilization of DNA polymerase by Sso7d. Sso7d, a tiny 7 kD protein, binds with the template DNA without any preference for specific sequences and stabilizes the association. This stabilization from the Sso7d fusion prevents the polymerase from dissociating during the process of amplification, increasing the processivity of the polymerase and thereby increasing the efficiency of a PCR/qPCR reaction. This is especially valuable when it comes to amplifying challenging sample types, such as those with PCR inhibitors, targets with high G-C rich regions, and longer amplicons.
Fusion protein technology has been extensively employed for tracking or visualizing biological processes. For example, visualization of proteins using the GFP fusion, expression of proteins using GST, and evaluation of protein interactions using yeast two-hybrid systems are all achieved using fusion proteins. Several strategies have been attempted to improve the processivity of the naturally existing DNA polymerases using the fusion protein technology, but none of these could be easily applied to thermally stable DNA polymerases widely used in PCR-based applications (Bedford et al. 1997, Davidson et al. 2003).
By far the best strategy demonstrating enhanced processivity and improved PCR efficiency is the approach developed and patented by Wang in 2000 and published in 2004 (Wang et al. 2004), where the nonreplicative DNA polymerase was covalently linked to the sequence-nonspecific dsDNA binding protein called Sso7d, obtained from Sulfolobus solfataricus (Figure 2).
Video. An animated description of the Sso7d fusion to DNA polymerase.
The Mechanism of Sso7d Binding to DNA Polymerase and How the Fusion Increases PCR Efficiency
Sso7d is a small protein (7 kD) capable of covalently binding to dsDNA without any preference for specific sequences. The binding of the Sso7d domain to a DNA polymerase, such as Taq polymerase, is optimal to smoothly slide along the template. The covalent linking of the fusion protein with the polymerase does not entail any structural modifications, therefore the fusion does not interfere with the structural integrity and thermal stability (Table 1), and consequently, with the catalytic activity of the enzyme. Sso7d can be linked to both family A and family B DNA polymerases and can bind with dsDNA at ambient temperature as well as high temperatures. It can therefore help enzymes that are thermostable or otherwise. Studies carried out using the Sso7d fusion polymerase have demonstrated that the fusion protein technique improves processivity without affecting the catalytic activity or thermal stability of the enzyme (Table 1).
Table 1. Improved processivity, catalytic activity, and thermal stability shown by Taq polymerase and Taq(Δ289), the mutant form of Taq, and a commercially available wild-type Pfu polymerase (adapted from Wang et al. 2004).
| Enzyme | Processivity (median number of nucleotides incorporated per binding event) | Turnover Rate (kcats-1) | t1/2 (min) at 97.5°C |
| Taq (Δ289) | 2.9 ± 0.7 | 18 ± 2 | 39 |
| Sso7d fusion to Taq (Δ289) | 51.0 ± 6 | 20 ± 1 | 35 |
| Taq | 22 ± 3 | 21 ± 1 | nd |
| Sso7d fusion to Taq | 104 ± 17 | 20 ± 1 | nd |
| Pfu | 6 ± 0.5 | 3.2 ± 0.1 | 730 |
| Sso7d fusion to Pfu | 55 ± 3 | 5.7 ± 0.2 | 789 |
Performance was measured in the full length Taq (which possesses an 5′-3′ exonuclease domain and a polymerase domain), Taq(Δ289) (which lacks the exonuclease domain and is considered to have less processivity than full length Taq), and Pfu ( a commercially available high-fidelity thermostable polymerase) with and without fusion with Sso7d protein. Processivity (assessed by median number of nucleotides incorporated in a binding event), catalytic activity (as determined by the turnover rate (kcat) of nucleotide incorporation at a saturating amount of dNTPs), and thermal stability (evaluated as half-life (t1/2) of the remaining activity of the enzyme after incubation at 97.5°C) were determined (see Wang et al. 2004 for further details). nd, not determined.
Fusion of a processivity factor to a polymerase is not guaranteed to provide effective nucleotide incorporation during PCR and not all fusion technologies increase PCR efficiency. For example, even though processivity factors such as thioredoxin or the clamp protein increase the processivity of DNA polymerase, they are not as thermally stable as Sso7d and exhibit less affinity toward dsDNA, rendering them impractical to use in PCR. The Sso7d protein binds directly to dsDNA with an affinity just strong enough to stabilize the polymerase and reduce the probability of dissociation of the polymerase from DNA, and just weak enough to enable the fusion protein to slide along the DNA template during nucleotide incorporation. The higher processivity afforded by the fusion polymerase helps reduce the extension time during PCR amplification and allows for the use of less enzyme. When both enzyme concentration and reaction times were considered, Wang et al. (2004) reported a 16-fold increase in efficiency in amplification of targets with longer amplicons when Sso7d was fused to the mutant Taq (Sso7d-Taq(Δ289) compared with no fusion (Taq(Δ289 ).
Fusion to Sso7d Enables Amplification of Longer Amplicons in Less Time
PCR efficiency can be negatively impacted when longer amplicons have to be amplified. It is imperative to have a polymerase with high processivity when amplifying longer DNA fragments. In addition, the time required for amplification of longer amplicons is also relatively high compared with shorter DNA fragments. Fusion with Sso7d confers the ability of the polymerase to amplify longer fragments in shorter times. Taq polymerase with Sso7d fusion (Sso7d-Taq(Δ289)) has been shown to amplify targets five times longer than can be amplified by its counterpart without Sso7d fusion (Taq(Δ289)). Similar enhancement of the ability to amplify longer targets with shorter extension times has also been confirmed with other commercially available polymerases such as Pfu (Wang et al. 2004).
Sso7d Fusion Imparts Greater Tolerance to PCR Inhibitors
A variety of impurities present in samples, especially crude and complex ones such as blood or soil, can inhibit PCR reactions. Inhibition by salt, in particular, could exert a destabilizing influence on the polymerase-DNA complex. But the Sso7d fusion is able to tolerate higher concentrations of salt than the Taq or Pfu polymerase without the fusion (Wang et al. 2004). Sso7d fusion has also shown superb tolerance to a myriad of other inhibitors (such as heparin, humic acid, indigo, tannic acid, cellulose, and trypsin), indicating that the fusion affords a unique advantage when targets with such PCR inhibitors need to be amplified.
Sso7d Minimizes PCR Bias during Preamplification
Sometimes the amount of sample available can limit the number of genes that can be analyzed by qPCR. The use of a preamplification step prior to qPCR is a common tool that expands the number of genes that can be analyzed for a given sample. A common fear around incorporating this step into the workflow is that it will introduce bias between genes and lead to inaccurate gene expression analysis. The heterogeneity among target genes can lead to the introduction of bias as some targets are amplified more easily than others. By preventing the enzyme from dissociating while amplifying difficult templates, the use of an Sso7d fusion polymerase ensures all targets are amplified with high efficiency, thus minimizing the bias that may be introduced during the preamplification step.
Sso7d Fusion Technology in the Development of PCR and qPCR Reagents
Sso7d fusion technology has been utilized in the development of PCR and qPCR reagents in order to reap the benefit of increased PCR efficiency. Bio-Rad Laboratories has a full line of PCR and qPCR products that employ Sso7d fusion technology. With the fusion polymerase and optimized buffers, these reagents provide extremely rapid hot-start activation and polymerization kinetics, facilitating fast PCR reactions. The fusion polymerase helps with amplification of long amplicons, samples with PCR inhibitors, and targets with high G-C content, increasing PCR efficiency in challenging amplification conditions. This increased PCR efficiency translates to more accurate and fast results, and increases the potential of PCR and qPCR to be used as a reliable tool for broader sample types and conditions.
![]() Commercialization of this technology has extended the capabilities of PCR and qPCR and increased our ability to obtain insights into questions that couldn’t be asked before.
Commercialization of this technology has extended the capabilities of PCR and qPCR and increased our ability to obtain insights into questions that couldn’t be asked before.
Some of the Bio-Rad reagents that incorporate the Sso7d fusion are: SsoAdvanced™ Universal SYBR®, SsoAdvanced Universal Probes, SsoAdvanced Universal Inhibitor-Tolerant SYBR®, and SsoAdvanced PreAmp Supermix, as well as iProof™ High-Fidelity DNA Polymerase.
References
Alvarez MJ et al. (2007). Model based analysis of real-time PCR data from DNA binding dye protocols. BMC Bioinformatics 8, 85.
Bedford E et al. (1997). The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA 94, 479–484.
Braithwaite DK and Ito J (1993). Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucelic Acids Res 21, 787–802.
Davidson JE et al. (2003). Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res 31, 4702–4709.
Ito J and Braithwaite DK (1991). Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res 19, 4045–4057.
Lalam N (2006). Estimation of the reaction efficiency in polymerase chain reaction. J Theor Biol 242, 947–953.
Laos R (2014). DNA polymerases engineered by directed evolution to incorporate non-standard nucleotides. Front Microbiol 5, 565.
Svec D et al. (2015). How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif 3, 9–16.
Wang Y (2004). A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res 32, 1197–1207.
Zhuang Z and Ai Y (2010). Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis. Biochim Biophys Acta 1804, 1081–1093.
SYBR is a trademark of Life Technologies Corporation. Bio-Rad Laboratories, Inc. is licensed by Life Technologies Corporation to sell reagents containing SYBR Green I for use in real-time PCR, for research purposes only.
Use of SsoAdvanced Supermixes is covered by one or more of the following U.S. patents and corresponding patent claims outside the U.S.: 5,804,375; 5,994,056; and 6,171,785. The purchase of these products includes a limited, non-transferable immunity from suit under the foregoing patent claims for using only this amount of product for the purchaser’s own internal research. No right under any other patent claim and no right to perform commercial services of any kind, including without limitation reporting the results of purchaser’s activities for a fee or other commercial consideration, is conveyed expressly, by implication, or by estoppel. These products are for research use only. Diagnostic uses under Roche patents require a separate license from Roche. Further information on purchasing licenses may be obtained from the Director of Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.