A Brooklyn Bridge too far
Danielle Joseph sat in the back of a slow-moving New York City cab. On the floor at her feet, a Styrofoam box held three vials of cells she had cultured for weeks, carefully prepared and stained, and packed on ice. She needed the cab to hurry up and get her — and those cells — over the Brooklyn Bridge.
Joseph was a graduate student at SUNY Downstate Medical Center, located deep in Brooklyn. Her research had reached a crucial point. She believed a key subpopulation of cells might help explain the poor treatment outcomes seen in multiple myeloma. She had some promising initial data, but her data meant nothing if she couldn’t confirm her experiments with higher quantities of cells. To isolate those cells, Joseph and her principal investigator, Dr. Stacy Blain, decided to reserve time on the cell sorter at New York University’s flow cytometry core lab.
See how the S3 Cell Sorter solved a major hurdle in Dr. Stacy Blain and Danielle Joseph’s multiple myeloma research.
Now, the many things that could go wrong on a cab ride — bridge traffic, construction, a broken water main, a driver’s bad route choice — stood between Joseph and the cell sorting she needed to move her project forward. “It was very stressful,” she remembers. “Because as you’re sitting there in traffic, every minute cells are dying. And if I didn’t have enough cells when I got back, I couldn’t complete the experiment.”
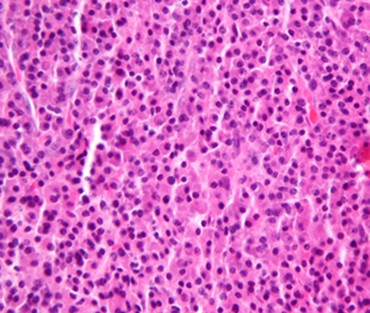
A microscopic view of malignant plasma cells, one of the clinical criteria for diagnosing multiple myeloma. Blain and Joseph want to find out if a small subpopulation of plasma precursor cells can remain quiescent and unaffected during chemotherapy, then reactivate and repopulate tumors. Image courtesy of Nephron, CC-BY-SA.
A mystery in multiple myeloma
Joseph’s research, the reason she needed a particular cell population in the first place, had its origin in a clinical question: Why is multiple myeloma frequently fatal? More specifically, why does disease progression in patients often follow a familiar and sad trajectory, first looking up before then turning down sharply?
“The most striking thing about multiple myeloma is the life expectancy,” Joseph says. “Patients who get the most advanced chemotherapeutic treatments live five, seven, sometimes ten years. They go into remission, even complete remission. But their tumor burden usually comes back and is then very often resistant to treatment.”
Multiple myeloma results from malignant plasma cells, which multiply and crowd the bone marrow, choking off the supply of other blood cells, weakening bones, impairing the immune system, and affecting organs such as the kidneys. The nature of today’s therapies, which show great initial success, may also point to the cause behind their ultimate failure, suggests Joseph’s principal investigator, Dr. Stacy Blain.
“Most of the current therapies that we have target rapidly proliferating cells,” Blain says. “However, we’re still not eradicating tumors. That suggests that there might be a cell that is resistant, evading the therapies. And so far we’re not targeting that cell.”
A stem cell for cancer
“Every cell in your body has to make this decision every day: should it divide or not divide?” Blain says. “My long-term interest is how cells regulate this decision.” While cancer is considered a disease of over-dividing cells, Blain, Joseph, and other researchers propose that a key subgroup of cancer cells doesn’t divide and instead remains dormant for years, dodging therapies that target cell proliferation, before suddenly reactivating and repopulating the tumor.
This is the cancer stem cell model, a relatively new hypothesis first advanced by leukemia and lymphoma researchers. By analyzing the malignant cells found in these disorders, scientists found a hierarchy of cell types similar to the hierarchy seen in healthy hematopoietic cells, where progenitor stem cells in the bone marrow create differentiated blood and immune cell progeny. A cancer stem cell, researchers proposed, might seed tumor tissue in much the same way.
“I wouldn’t say I came into this [project] believing in cancer stem cells,” says Blain. “I didn’t see the need to evoke a whole new model to explain the [multiple myeloma] tumor dormancy phenotype. My evolution in thinking occurred in the best possible way: by Danielle showing me results and me having to scratch my head and say, ‘wow that’s really interesting and awesome.’”
Crisis in the incubator
Back in Danielle Joseph’s taxi, things weren’t going well. On the days she went to NYU to sort cells, she left Downstate Medical Center early and returned late, rushing to get the sorted cells out of their buffer solution and back into the incubator. However, mornings after these trips usually found her and Dr. Blain not moving forward with research but instead surveying the damage.
“I’d ask, ‘How are we doing?’” Blain remembers. “And Danielle would say, ‘Well, 25 percent of the cells are still alive.’ 25 percent! As her PI who was paying for this enterprise, that was really distressing.”
In investigating multiple myeloma cancer stem cells, using the RPMI cell line, Joseph homed in on cells lacking the surface marker CD138 as the likely cancer stem cell. “CD138 and CD38 are well-known markers for plasma cells,” Joseph says. “CD138 is specifically the marker that distinguishes between a precursor plasma cell and a fully differentiated plasma cell.”
For two years, she isolated the CD138-negative population and in the process mastered the one archaic cell sorting instrument at Downstate Medical Center, calculating drop delay by counting beads on a microscope slide while carefully realigning the laser throughout the sorts, and painstakingly produced enough data to justify the trips to NYU’s core lab. Despite the frustration and late nights, Joseph grew to love the technique as she learned its ins and outs.

PhD candidate Danielle Joseph (foreground) and Stacy Blain (background), Assistant Professor at SUNY Downstate Medical Center, stain cells from the RPMI cell line with antibodies for CD138 in preparation for sorting them using the S3 Cell Sorter.
In various assays, the CD138-negative population showed key characteristics of cancer stem cells: quiescence, self-renewal, the ability to grow into differentiated progeny, and chemoresistance. She needed the NYU sorts to give her more cells to validate her studies and further investigate signaling mechanisms, so to have so many of those cells die in a taxi was a major setback.
At that point, Joseph’s project was in jeopardy, as Blain remembers: “Here I am, I’m a mentor of a graduate student who’s already invested a couple years, she’s made great strides on the project. But, realistically we’re not going to be able to continue it unless we win the lottery. That would have been a devastating conversation to have with Danielle.”

The S3 Cell Sorter is a benchtop instrument that gives individual researchers like Blain and Joseph the ability to sort up to four colors at a time without requiring a trained technician. Most instrument functions are automated and controlled through a user-friendly software interface, allowing a wide range of students and investigators to sort cells.
Little sorter, big difference
Instead of shutting down Joseph’s project, Blain found a solution by switching to a cell sorter that could do basic cell sorting right in her lab. The instrument she acquired was the Bio-Rad S3 Cell Sorter. In doing so, she wanted to both support Joseph’s research and lay the groundwork for similar experiments by other students. “I want to be able to say ‘yes’ to my students,” she says. “I want to be able to say, ‘you thought of a great idea, let’s try it.’”
The S3 Cell Sorter is a compact and simple cell sorter that automates the sorting process through an array of systems: ProDrop™ technology to set drop delay, the AutoGimbal™ system to align the nozzle tip and sort stream, as well as various quality control systems to detect and correct bubbles and other potential problems. These systems are run by the instrument’s ProSort™ software, so that, other than collecting vials at the end of a sort, users interact with the instrument entirely through an intuitive desktop interface.
“No longer did I have to align the laser or set the drop delay,” says Joseph, recalling the highly technical manual tasks of sorting that the S3 Cell Sorter automates. “I wasn’t used to working with a cell sorter that was that compact but still had the precision and accuracy that [larger] sorting flow cytometers had.”
Having the S3 Cell Sorter on-site also significantly reduced the amount of time Joseph needed to get CD138-negative cells. From a starting sample of 10 million cells, Joseph estimates that she got about 100,000 viable CD138-negative cells after a long day spent going to NYU’s core lab, then returning to SUNY Downstate Medical Center. Now, she says, “I can get 100,000 [CD138 negative] cells in maybe 10 minutes on the S3.”
With the new cell sorter up and running, Joseph found the pace of her research finally picking up, both in terms of sorting and in downstream experiments. “When I was going to NYU, I was doing one experiment a week,” she says. “That was really slow progress. Now I can sort every day, so that’s almost 10 experiments every week. Now my problem is trying to manage my data and analyze it so I can present it.”
Passing the purity test
Population integrity had been another major issue up to this point. First, when Joseph had used her department’s old sorter, the constant adjustments it required resulted in varying purity levels with each sort — 77% in one, 98% in another, 85% in the next. Later, in cabbing to the east side of Manhattan to sort at NYU, the high amount of stress her cells endured during the trips, and the high percentage that died, led to doubts about the cells that did survive: Were they a representative sample?


Joseph holds cells from her RPMI multiple myeloma cell line (left) suspended in media before preparing the cells for sorting. After staining and diluting the cells in suspension, she loads them (right) into the S3 Cell Sorter.
“If we’re trying to compare CD138-negative and CD138-positive populations, it’s very important that we’re starting with those populations, not mostly CD138 negative and mostly CD138 positive,” Joseph says. “I really appreciate the S3 [Cell Sorter] for allowing us to be confident that we’re getting reliable data on these groups.”
From her perspective as Joseph’s PI, Blain sees this reliability as a timesaver as well. “Danielle and I used to spend so much time looking at dot plots, wondering if we had obtained a pure population,” she says. “With the S3 [Cell Sorter], she never shows me those any more. I know she can say, ‘Yup, 99% pure’ every time. That’s a big difference. We’re doing important experiments and not worrying about the process so much.”
Ready to defend their data
One challenge Joseph had with the RPMI cell line was that her population of interest, the CD138-negative cells, represented only 5% of the total population. Having such a small population and only limited resources to sort it had produced a bottleneck, especially since the true focus of Joseph’s work wasn’t on isolating the cells, but in characterizing their behavior once she could observe them as a pure population.
“We’re putting the isolated [CD138-negative] cells back into culture,” she says. “We want to look at DNA levels to see if the cells are quiescent or rapidly going into the cell cycle. We’re also looking at cytokine release in the different populations. Being able to do our sorts at a higher rate also speeds up all the assays we’re doing after.”
Having done enough studies to make their findings statistically significant, now Joseph and Blain are preparing to publish their work. They are excited to both bolster the evidence that the CD138-negative population Joseph investigated is in fact the multiple myeloma cancer stem cell, and to reveal a novel finding about what occurs after cell differentiation. This finding may shake up the cancer stem cell model some, but Joseph feels ready for any controversy. “We can have confidence in defending our story because we can say, ‘we started with a pure population and this is what we saw,’” she says.

Blain and Joseph review cell differentiation assay data, which represents one criteria for assessing a possible cancer stem cell population. Because the most important assays in Blain and Joseph’s research occur after cell sorting has taken place, it’s essential that those sorts result in pure populations of CD138-positive and CD138-negative cells.
Focused on science, not sorting
Cell sorting is now an everyday technique in Blain’s lab, a major change she’s noticed over the course of her research career. “When I was in grad school, doing flow cytometry was the experiment,” says Blain. “It was earth-shattering if you could find a small population and isolate it. Now [with the S3 Cell Sorter] we are focusing on the questions we’re asking after we sort. We’re focusing on the science, not the sorting.”
At a time when funding for biomedical research continues to decline, Blain points to the improvement in cell sorting in her lab as leveling the playing field for her. “Science should be limited not by my resources but by my mind,” she says. “No one knows where the next great idea is going to come from. With the S3, I don’t have to be limited by a big expensive technique. I can say yes more to my students. It feels very different to be a mentor who can say that.”
With Joseph preparing her work for publication and herself for graduation, other students in the lab are building on her work. Priyank Patel, a newer grad student in Blain’s lab, recently started a similar series of investigations into cancer stem cells using a breast cancer line. One of the first steps he completed in this effort was successfully sorting out his population of interest on the S3 Cell Sorter.

PhD candidate Priyank Patel, Danielle Joseph, and Stacy Blain celebrate a successful first sort. Patel wants to investigate the cancer stem cell hypothesis in breast cancer. Being able to sort the subpopulation he will study represents a first step in this effort.

