One-step multiplex RT-qPCR is a technique used to rapidly quantify multiple targets directly from RNA in a single reaction.
Advantages of one-step multiplex RT‑qPCR:
- More data from each sample
- Increased throughput
- More accurate data normalization
- Reduced pipetting
- Cost effective
Common applications of multiplex PCR:
- Gene expression analysis
- Virus/pathogen detection
- Diagnostic research
- Small molecule screening
- Drug discovery
- Dosage experiments
- CRISPR screening
- Toxicology studies
Multiplex RT-qPCR Assay Design and Optimization
Proper optimization and validation is essential for successful one-step multiplex RT-qPCR. This initial investment of time up front ensures that resources and samples are not wasted producing suboptimal data later on. Optimization is particularly important for labs that repeatedly analyze the same targets, where the assay conditions will be used for many downstream experiments.
- All primers should have a similar Tm, 55–60°C
- Each probe’s Tm should be 8–10°C higher than the Tm of the primers
- Target amplicons should be 70–150 bp for optimal qPCR efficiency
- The primers should be screened to minimize the likelihood of primer-dimer formation
- Follow other general PCR primer design rules
Tips:
- It is important to make sure that the different primer and probe sets do not exhibit homology to one another. Remember that all primers and probes will be present in a single reaction
- Use commercially available software or open source design tools to design optimal primer-probe sets for multiplex assays. Primer3, Primer3Plus, and Primer-BLAST are good examples of open source tools
- Some oligo manufacturers also offer primer and probe design tools
- Consider using predesigned primers and probes to streamline this first step. PrimePCR Probe Assays for Gene Expression are available for multiplex detection of known gene targets
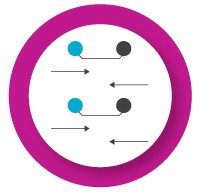
- Select fluorophores that are compatible with your real-time instrument. Check your real-time instrument manual for a list of supported fluorophores. Some common real-time instruments can be calibrated to use fluorophores that are not included in the default settings
- Select reporter fluorophores with minimal overlap in their emission spectra to reduce signal crosstalk between channels
- Consider total fluorescence signal intensity — apply the brightest fluorophores to the probes for the least abundant targets. This minimizes the opportunity for signal crosstalk between channels and can improve signal:noise ratio
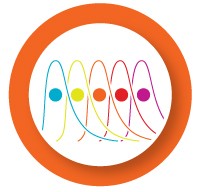
Before starting a multiplex reaction, it is important to test the dynamic range and reaction efficiency of each assay in singleplex. To do so, generate a standard curve using a template dilution series for each primer/probe set.
Considerations for reaction setup:
- Use the same sample type (purified RNA, DNase-treated RNA, lysates, or other sample types) and the same reagent to be used in the multiplex reaction
- Adjust the primer-to-probe ratio for optimal results based on expression levels, starting at a standard ratio of 2:1. Increase the ratio for low-abundance targets. A ratio of 1:1 is recommended for highly abundant targets
Acceptable performance criteria:
- The efficiency of the individual reactions is 90–105%
- The R2 of the standard curve is > 0.980, or r > |–0.990|
If assay performance does not meet these criteria, optimize the individual assays as you would for conventional qPCR. For tips, see our guide to qPCR best practices.
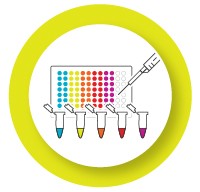
Validated individual assays are ready to be put together into a multiplex reaction. Validate the multiplex reaction to ensure that the dynamic range and reaction efficiency remain the same.
- Run singleplex and multiplex reactions for your targets on the same plate
- Calculate the ΔCq between the singleplex and multiplex reaction for each target
(ΔCq = Cqmultiplex – Cqsingleplex) - A ΔCq less than 1 indicates no significant change in reaction efficiency between the singleplex and multiplex reactions. Congratulations! You can proceed to running the multiplex assay on your samples
- A ΔCq greater than 1 indicates significant change between the singleplex and multiplex reactions. Additional optimization is needed (go to step 5)

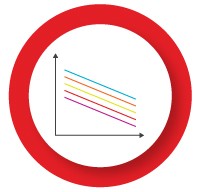
The individual assays in the multiplex reaction can compete with each other for reaction components, such as DNA polymerase and dNTPs, which may become limiting in later PCR cycles. This may result in poor detection of targets which are less abundant or not efficiently amplified. Optimizing the concentration of key reaction components can help address this problem:
- Decrease the primer concentration for the highly abundant target — leads to primer depletion and an earlier plateau
- Add more DNA polymerase — a multiplex reaction generally requires at least twice as much enzyme as a singleplex reaction
- Add more dNTPs — amplification of multiple targets in the same reaction requires more dNTPs
- Add more Mg2+ — the increased quantity of RNA, primers, probes, and dNTPs in a multiplex reaction decreases the availability of Mg2+ in solution
Reagent Selection for One-Step RT-qPCR
Several manufacturers offer RT-qPCR supermixes designed specifically for multiplexing, which may reduce the time required for optimization. Consider the following as you choose the right reagents for your needs:
- Multiplexing capability — assessing more targets per reaction saves time, reagents, and sample
- Sample type compatibility — tolerance to PCR inhibitors in lysates or samples of varying quality
- Detection sensitivity — linear range of detection varies by reagent
- Sample input range — choose a product suitable for your sample and targets
Bio-Rad’s Tools for Painless Multiplex One-Step RT-qPCR
Reliance One-Step Multiplex RT-qPCR Supermix — the best choice for researchers who want to reliably and reproducibly quantify multiple RNA targets directly.

For more information on Reliance One-Step Multiplex Supermix, check out the Bio-Rad website and our related articles.
PrimePCR Probe Assays for Gene Expression — create your custom multiplex assay pool with ease.
- Hundreds of predesigned and validated assays for 12+ common organisms
- Probes designed with multiplexing in mind, for maximum specificity and minimal cross-reactivity
- Available with FAM, HEX, TEX615, Cy5, or Cy5.5 fluorophores
For more information on PrimePCR Assays, check out the Bio-Rad website and our related articles.

