Dr John J Rossi’s titles and accolades are many and varied — and well earned. In his current affiliation with the City of Hope Comprehensive Cancer Center, Rossi serves as chair and professor of molecular and cellular biology, dean of the graduate school of biological sciences, and associate director for laboratory research. He is co-leader of the cancer biology program and the first holder of the Lidow Family Research Chair. These professional accomplishments are complemented by numerous awards, including a 2002 Merit Award in the Division of AIDS, National Institute of Allergy and Infectious Diseases. The common thread that weaves all of these activities and achievements together continues to be an unabashed enthusiasm for and curiosity toward scientific discovery — specifically in the molecular genetics of disease.
Rossi received his doctoral degree in microbial genetics in the late 1970s. At the time, cloning was only just becoming a tool that researchers could use, and with Rossi’s exposure to this now basic technique, his fascination with genetics turned to the molecular aspects of the discipline. Rossi was drawn to postdoctoral studies in Dr Arthur Landy’s lab at Brown University because of Landy’s groundbreaking work in sequencing genetic information for the bacteriophage lambda. Landy’s work focused on trying to understand some of the sequences of the attachment site of the bacteriophage in its host chromosome. He also completed the first restriction map of any lambda phage. Rossi was particularly attracted by the technology he would have access to in this forward-thinking environment.

In the early 1980s, Dr Keiichi Itakura with the City of Hope, whom Rossi refers to as “one of the fathers of DNA synthesis,” was looking for a person to work with synthetic DNA and apply it to some problems in biology and medicine. “Today, it’s a routine tool; you don’t even think twice about ordering a piece of DNA,” says Rossi. “But at the time, it was a hot area that was just evolving, so I came here [to the City of Hope] initially to learn how to make DNA, and then how to use it.”
Rossi’s early projects with the City of Hope focused on developing antisense technology. “Because we had synthetic DNA capabilities, we wanted to see if we could use it to block gene function,” explains Rossi. “We probably had the first example of using an antisense oligonucleotide to block replication of a virus.” But because the research was conducted in bacteria and not considered publication worthy, his group never submitted the findings. This project did, however, lay the foundation for similar studies related to ribozymes. “Ribozymes have the capability of cleaving the target — this became fascinating because I wanted to understand more about the mechanisms and how they would function inside a living cell,” Rossi explains. His research focus continued primarily on ribozymes — until RNA interference (RNAi) was discovered.
Rossi considers the discovery of RNAi as pivotal to antiviral discovery. “RNAi has become another important, powerful genetic tool we can use to further our antiviral work,” he says. Rossi’s lab became the first to publish research demonstrating the expression of small interfering RNAs (siRNAs) to inhibit HIV replication in human cells. Various aspects of RNAi and siRNAs continue to play a key role in his research on viral diseases and lymphomas.
One of the stumbling blocks inhibiting progress in his latest research efforts, however, has occurred in the transfection step of the RNAi workflow. “When studying siRNAs, you have to get the synthetic molecules into the host cells, and for many cultured cells — especially if they’re adherent to the place in which they’re grown — it’s common to use lipid-based transfection agents. But for primary cells such as B cells, T cells — they’re impossible to transfect,” Rossi explains. This is where electroporation comes into play.
Currently, Rossi’s group is seeking the genetic mechanisms critical to the suppression of B-cell lymphoma proliferation. Lymphomas are cancers derived from lymphocytes, a type of white blood cell that protects the organism from foreign bodies such as bacteria and viruses. B-cell lymphoma is caused primarily by chromosomal translocations and constitutes approximately 85% of all non-Hodgkin’s lymphomas. In a recent experiment, his group introduced bifunctional siRNAs (each strand targets different genes) that target suspected anti-growth pathways into various B-cell lines to effectively knock out the ability of these cells to proliferate (Figure 1).
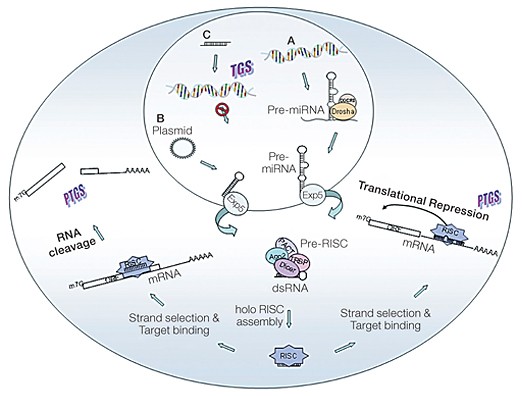
Fig. 1. Mechanism of cellular gene silencing. A, Primary microRNAs (pri-miRNAs) are processed by Drosha and its partner DGCR8 into precursor miRNAs (pre-miRNAs) and transported to the cytoplasm by exportin 5 (Exp5). In the cytoplasm, they are processed to yield the guide sequence containing all the components required for gene silencing. The guide sequence binds to the target sequences in the 3′ UTRs. If the miRNA guide sequence is fully complementary to its target (left pathway), site-specific cleavage and degradation of the mRNA is triggered. If the base pairing is incomplete (right pathway) but includes pairing of the seed region with the target, translational inhibition occurs, often accompanied by messenger degradation. B, Artificially transcribed shRNAs are transported to the cytoplasm in a way similar to that of miRNAs. The dsRNA in the cytoplasm is recognized and processed by Dicer into ~21–25 nucleotide siRNA fragments that are loaded into the RISC. The siRNAs target complementary sequences of cellular mRNAs and trigger their degradation through AGO2-mediated cleavage. C, When siRNAs are present in the nucleus and are complementary to promoter regions, they can trigger transcriptional gene silencing (TGS).
The cells used in Rossi’s experiments were all in suspension, so each line was different, and determining electroporation conditions became a major obstacle to pursuing this avenue of experimentation. “We tried the amaxa electroporator, ventromers — you name it and it seems like we tried it,” says Rossi. But nothing enabled them to electroporate siRNAs into B cells and retain cell viability — until his lab was offered the opportunity to work with Bio-Rad’s Gene Pulser MXcell™ electroporation system (Figure 2).
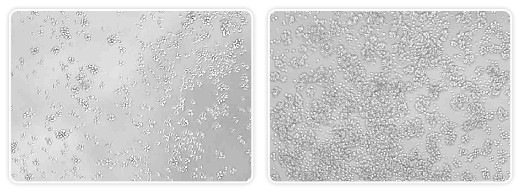
Fig. 2. Bright field image of human B cells. Jeko (left) and Z138 (right) cells were transiently transfected with siRNAs using the Gene Pulser MXcell electroporation system. Cells were viewed 24 hr posttransfection under an inverted microscope using bright field imaging. Other attempts (amaxa electroporator and lipid transfection) to transfect into these cell lines were unsuccessful.
The Gene Pulser MXcell system is a flexible platform allowing both manual and preset programming. Because the Gene Pulser MXcell system allowed Rossi’s group to perform multiwell analyses and test multiple conditions simultaneously, they were finally able to determine conditions in which “the cells weren’t dying, we were getting transfection, and we could monitor target knockdown,” says Rossi. “The Gene Pulser MXcell system proved very, very useful — I think it’s a great little invention.”
Ultimately, the group was able to demonstrate that the bifunctional siRNAs of interest can independently knock out their targets in B cells (data pending publication).
As it was in the beginning of his career with the fascination of cloning as a tool to uncover genetic mysteries, Rossi today remains intrigued by the application of cutting-edge tools and technologies to help further research.
For more information, request Bulletin 5942.

