Genetic screens are powerful tools for finding genes associated with a particular phenotype. They play an especially important role in cancer research and are leveraged to identify predictive biomarkers that can be used to improve treatment efficacy or trace the progression of genetic alterations that ultimately lead to disease. The HAP1 cancer cell line model and fluorescence-activated cell sorting (FACS) bring efficiency to the development of genetic screens that is making them a force to be reckoned with.
The Challenge with Genetic Screens: The Two-Allele Problem
The diploid nature of human cells makes it difficult to link genotype to phenotype. Except for gametes, all cells in sexual organisms are diploid, carrying two copies of each gene. A genetic mutation in one copy can be compensated for by the other. This has implications for genetic screens; a genetic mutation may not be reflected in the organism’s phenotype due to genetic redundancy.
Haploid cells, by contrast, cannot mask the effect of mutagenesis and are used extensively for random mutagenesis screens (Li and Shuai 2017). Using haploid human cells such as the HAP1 cancer cell line, a genetic mutation can be linked to phenotype within a cell. CRISPR-Cas9 mutagenesis has become a popular method for forward genetic screens (Figure 1).
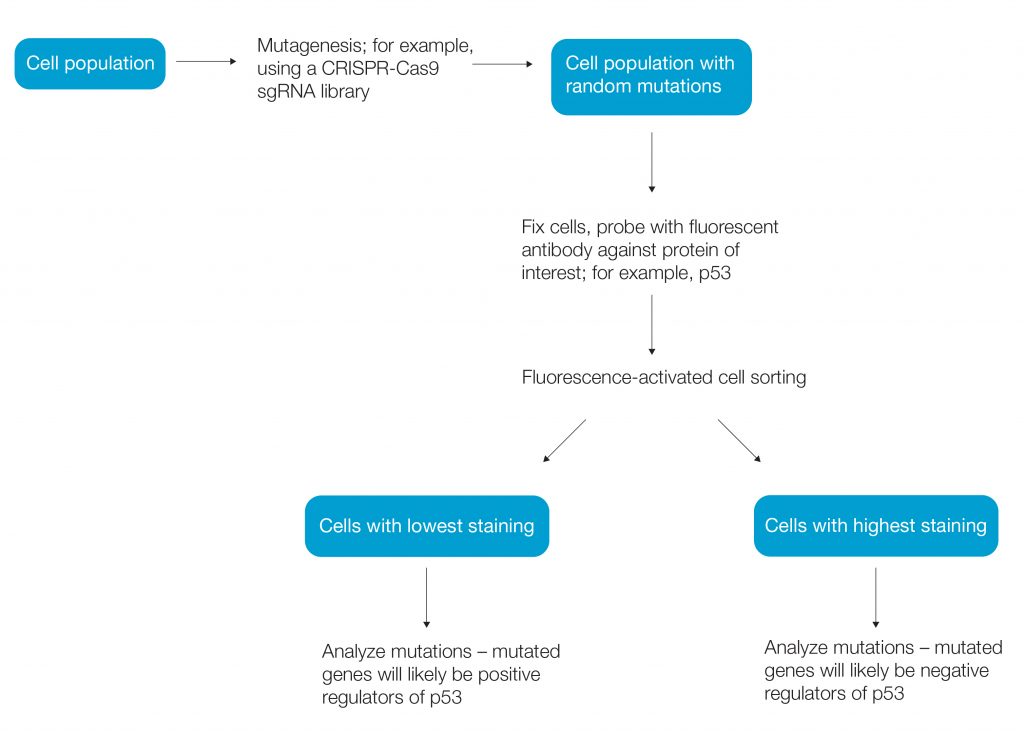
Fig. 1. Workflow of a simple genetic screen.
HAP1 Genetic Screens in Cancer Research
Studies have used HAP1 genetic screens to identify genes involved with protein regulation. A 2018 study by Alsamman and El-Masry, for example, found that interferon regulatory factor 1 (IRF1) controls cell proliferation and survival, contributing to tumor biology. Understanding IRF1 regulation is important to cancer research.
Researchers at the Netherlands Cancer Institute used random mutagenesis in HAP1 cells and then used antibody staining to sort cells based on levels of IRF1, expression of which is induced by interferon-γ (IFN-γ) under strong genetic control. By comparing genetic mutations in the high- and low-IRF1 populations, they were able to identify genes involved with IRF1 production or regulation (Brockmann et al. 2017).
Genetic screens can identify proteins that regulate protein-protein interactions. CD47 provides a “don’t eat me” signal to macrophages and is overexpressed in cancer, contributing to evasion of the immune system. Per Willingham et al. (2012), its interactions are with SIRPα on macrophages and dendritic cells. To identify regulators of CD47-SIRPα binding, Logtenberg et al. (2019) performed a FACS-based haploid genetic screen using an antibody that binds to the SIRPα recognition site.
Mechanisms of resistance to anticancer therapies can be studied using genetic screens. Gerhards et al. (2018) identified genes whose loss creates sensitivity to a class of anticancer drugs called microtubule-targeting agents (MTAs). They treated mutagenized cells with docetaxel or vinorelbine and then identified, sorted, and sequenced surviving cells. Genetic mutations gained by surviving cells gave insight into genes involved in drug resistance.
The goal of precision medicine is to guide genotype-specific treatment. Using a genetic screen, Mezzadra et al. (2017) aimed to identify proteins that regulate PD-L1, which could identify patients who would respond to PD-1 or PD-L1 antibody treatment as cancer therapy. Researchers screened for modulators of PD-L1 in HAP1 cells that had been treated with IFN-γ to induce PD-L1 expression. They fixed and stained cells and sorted them into populations with the highest and lowest PD-L1 expression. They then isolated genomic DNA from the sorted cells and identified the mutated sites.
Why FACS for Genetic Screens
One of the major challenges with some types of genetic screens is separating out the relatively rare events (valid hits) from high numbers of false positives. Biology is never black and white and, depending on your phenotypic readout, any increase or decrease in expression level may be minimal, making it hard to separate out the noise. Other factors, such as gene knockout, knockdown efficiency, or library size, can further negatively affect the signal-to-noise ratio, making it even more difficult to separate out true positives. Low event frequency, abundant false positives, and poor mutagenesis efficiency is why large numbers of cells are required for genetic screens (up to 100 million for CRISPR-Cas9 knockout screens).
One way to improve the likelihood of isolating a true hit is to use a selection method that is both sensitive and very high throughput, such as FACS. Genetic screens often involve a simple cell sort for intracellular levels of a certain protein tagged by a fluorescent antibody because this method maintains sample purity while allowing rapid and easy selection of hits based on fluorescence levels. Specifically, the sensitivity of cell sorting makes it easy to isolate the top 1–5% of hits, which helps ensure isolation of valid hits and minimization of false positives. With a sorting rate of 10,000 cells per second, it is possible to screen large numbers of cells in a very short time.
The Cell Sorter Anyone Can Use
Cancer research studies have benefited greatly from incorporating FACS into their genetic screen workflows. But not all researchers are comfortable with cell sorting and not all cell sorters are designed with novice users in mind. The S3e Cell Sorter not only provides the sensitivity, speed, and purity essential for robust genetic screens, but it is also suitable for use by those new to flow cytometry. With automated and turnkey setup, intuitive instrument controls that guide the user through the cell sorting process, and optics ideal for sorting cells expressing fluorescent proteins or labelled with commonly used dyes, the S3e Cell Sorter is truly the cell sorter that anyone can use.
Click here for more information about the S3e Cell Sorter.
References
Alsamman K and El-Masry OS (2018). Interferon regulatory factor 1 inactivation in human cancer. Biosci Rep 38; doi: 10.1042/BSR20171672.
Brockmann M et al. (2017). Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature 546, 307–311.
Gerhards NM et al. (2018). Haploid genetic screens identify genetic vulnerabilities to microtubule‐targeting agents. Mol Oncol 12, 953–971.
Li Y and Shuai L (2017). A versatile genetic tool: haploid cells. Stem Cell Res Ther 8, 197.
Logtenberg MEW et al. (2019). Glutaminyl cyclase is an enzymatic modifier of the CD47-SIRPα axis and a target for cancer immunotherapy. Nat Med 25, 612–619.
Mezzadra R et al. (2017). Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110.
Willingham SB et al. (2012). The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A (PNAS) 109; 6662–6667.

