Fluorescence-activated cell sorting is a valuable tool for research, allowing separation of a mixture of cells into distinct populations for analysis. But could the physical forces of sorting stress the cells, altering their behavior and confounding experimental results? Andrä et al. (2020) found that although cell sorting activated p38 MAPK stress signaling, no functional or structural changes were observable. The results indicate that jet-in-air systems such as the Bio‑Rad S3e Cell Sorter do not alter cell behavior.
Fluorescence-Activated Cell Sorting Has a Wide Range of Uses
Like regular flow cytometers, cell sorters analyze cells based on a fluorescent component such as a fluorescent antibody bound to a target of interest. While regular flow cytometers evaluate fluorescence properties, cell sorters physically separate cells based on those properties. Distinct populations of cells are collected in separate vials. Researchers can then use them for further experimentation by comparing sorted populations, or by studying one population.
Fluorescence-activated cell sorting is widely used in a variety of research applications. For example, rare cell populations can be isolated by cell sorting and their gene expression profiles discovered using single-cell RNA sequencing (RNA-Seq) (Figure 1).
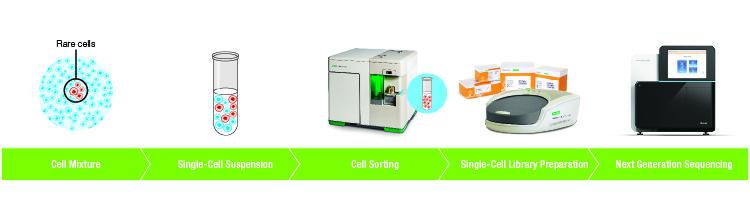
Fig. 1. Example workflow of cell sorting upstream of single-cell sequencing.
Cell Sorting Exposes Cells to Physical Forces
Cells experience a range of forces during cell sorting that can cause stress and confound downstream results.
Andrä et al. examined the forces that cells undergo during sorting and assessed the effects on cellular responses. They used the Bio‑Rad S3e Cell Sorter, among other instruments, to study activation of p38 MAPK stress signaling and functional indicators of cell stress. Human peripheral blood mononuclear cells (PBMCs), murine splenocytes, and immortalized cells were studied. The investigators compared stress indicators of different cellular populations, including mock sorted cells. Mock sorted cells not treated with fluorescent antibodies were passed through a flow cytometer and sorted on scatter properties in order to isolate the mechanical and physical forces of cell sorting from the effects of antibody staining. The mock sorted cells were then compared with unsorted cells to assess the effects of mechanical and physical force.
Cell Sorting Was Found to Activate p38 MAPK
Comparing mock sorted versus unsorted PBMCs, researchers found that cell sorting activated p38 MAPK by shear forces or acceleration. Oscillation, laser light, electric charge, and impact caused no further activation. The study found that p38 MAPK was activated at all temperatures, and no variation of sorting conditions prevented activation.
Sorting Did Not Affect Cell Function or Structure
Cell survival is affected by p38 MAPK activation, so the researchers looked at viability. Cell survival was found to be similar in sorted and unsorted cell populations. The ability of T cells to perform antigen-specific killing is also linked to the p38 MAPK pathway. This function was not significantly impeded by cell sorting, although the researchers did note that primary CD8+ T cells might not be the most sensitive cell type to sorter-induced or environmental changes.
As p38 MAPK is a regulator of the cell cycle, the scientists investigated cell cycle progression after sorting. Activation of p38 MAPK did not result in cell cycle arrest in sorted versus unsorted cells. Migratory behavior of CD8+ T cells was also not found to be impaired by sorting. No structural differences and no deformation or ruptures in the cytoskeleton were observed in sorted versus unsorted cells.
Gene Expression Was Not Significantly Affected by Cell Sorting
As a protein kinase, p38 MAPK is capable of activating transcription factors within many signaling pathways, it seemed likely that p38 MAPK activation could modify gene expression profiles in sorted cells. However, researchers found that p38 MAPK phosphorylation induced little or no change in mRNA expression patterns.
This is a welcome finding for anyone analyzing gene expression of cell populations after sorting, whether by RNA-Seq or other methods.
Findings Suggest That the S3e Cell Sorter Is Gentle on Cells
The experiments found that although cells responded to the external stresses of sorting, there were no meaningful changes to behavior or physiology. These results could help build confidence in fluorescence-activated cell sorting, with a device such as the S3e Cell Sorter, as a technique when downstream experiments on the sorted cells are needed. For example, during the isolation of low-frequency or rare cell populations followed by RNA-Seq in an experiment performed by Bio‑Rad (Figure 2).
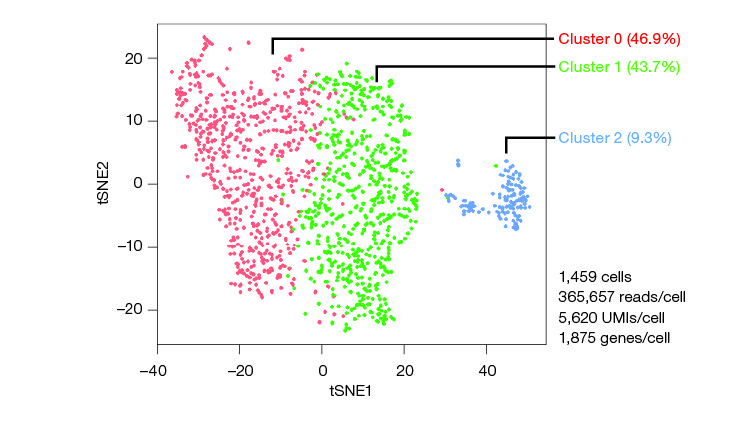
Fig. 2. CD34+/CD45+ human hematopoietic stem and progenitor cells are among the rarest subpopulations of PBMCs. In an experiment performed by Bio‑Rad, purified PBMCs were stained using FITC anti-human CD45 and PE anti-human CD34 and sorted using the S3e Cell Sorter. Transcriptomes of isolated CD34+/CD45+ cells were sequenced using the Illumina® Bio-Rad Single-Cell Sequencing Solution. Unbiased clustering was performed. Analysis revealed at least three transcriptionally distinct populations (clusters 0, 1, and 2). UMI, unique molecular identifier.
The S3e Cell Sorter is easy to use, requiring minimal training to operate. It can be left running while a user focuses on other tasks, maximizing efficiency. The S3e Cell Sorter is fast but the high sorting speeds do not compromise sample purity or cell integrity.
Compared with magnetic-activated cell sorting (MACS) or other bead-based approaches, the S3e Cell Sorter offers greater flexibility in marker selection. When used in combination with single-cell sequencing, the S3e Cell Sorter improves data quality by eliminating background RNA and reduces sequencing costs by ensuring that a homogeneous sample is free of dead cells and debris prior to being sequenced.
Findings from Andrä et al. suggest that fluorescence-activated cell sorting can be gentle on cells. This is positive news for anyone using systems such as the S3e Cell Sorter upstream of single-cell analysis or other experiments.
References
Andrä I et al. (2020). An Evaluation of T‐Cell Functionality After Flow Cytometry Sorting Revealed p38 MAPK Activation. Cytometry A 97, 171–183.
Watson A et al. (2018). Identifying Heterogeneity within Rare Cell Populations by Pairing Single-Cell RNA-Seq with Cell Sorting. Bio‑Rad Bulletin 7055.
Bibliography
Zarubin T and Han J (2005). Activation and signaling of the p38 MAP kinase pathway. Cell Res 15, 11–18.

