Surface Plasmon Resonance (SPR) is an optical phenomenon that is used for the label-free analysis of the binding of any two molecules in real time. An SPR biosensor instrument generates a sensorgram by monitoring the change in the SPR response with time. Fitting is possible with an instrument-generated sensorgram, and fitting the sensorgram to a suitable kinetic model allows for the calculation of kinetic parameters such as the association (ka) and dissociation (kd) rate constants. SPR is an important tool for characterizing biomolecular interactions.
Although SPR is widely used in biomolecular interaction analysis, it has some drawbacks, such as the inability to deliver specific molecular weights of analytes, thus resulting in uncertainty with respect to the specific species being measured. The coupling of SPR and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) overcomes this disadvantage. In traditional SPR molecules are collected by the elution of analytes from the sensor chip surface after kinetic analysis. However, since the SPR sensor chips are coated with a conductive gold layer, they are highly adaptable to MALDI-TOF-MS, which allows for direct detection of surface captured proteins on an SPR sensor chip with no elution steps. Compared to the traditional method, direct detection from the sensor chip surface could lead to enhanced sensitivity and a simplified sample preparation process.
The ProteOn XPR36 protein interaction array systemis an SPR platform that features a 6 x 6 multiplex experiment configuration for simultaneous analysis of multiple targets and analytes. This unique patented design facilitates running 36 different interactions in a single analyte injection, which is known as the One-shot Kinetics™ approach. With this unique design, the ProteOn XPR36 system allows for high data quality, versatility in experimental design, and high-throughput sample processing.
We describe in this article the method of combining SPR and MALDI-TOF-MS for characterizing biomolecules after kinetic analysis. Results of an experiment conducted as a proof of principle by running protein-protein interaction analysis in the ProteOn XPR36 system, immediately followed by direct MALDI-TOF-MS analysis of the protein analyte on the same sensor chip, are presented below. In this workflow, kinetic analysis is achieved in the first step, followed by molecular weight characterization in the second step. The molecules used in this study were β-amyloid 1–40 fragment and an anti-β-amyloid 6E10 monoclonal antibody.
Kinetics Analysis Using SPR
The SPR experiment was carried out on a ProteOn GLH sensor chip. 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysulfosuccinimide (sulfo-NHS) were sequentially injected in the ProteOn XPR36 system to activate the carboxyl groups on the sensor chip surface. Following that, β-amyloid 6E10 monoclonal antibody diluted to 25 µg/ml in pH 4.5 acetate buffer was injected to immobilize the antibody. Ethanolamine HCl was used to deactivate the sensor chip surface after the immobilization. The β-amyloid 1–40 fragment analyte stock solution was prepared in either phosphate-buffered saline with high (0.05%) Tween 20 (PBSHT) or PBSHT with 10% human serum, and diluted in the running buffer to make a dilution series for kinetic analysis. Two separate GLH sensor chips were used to screen these two differently prepared analyte samples. The running conditions for the two sensor chips were the same, except that PBSHT with 1% BSA was used as the running buffer for the serum-containing analyte sample while PBSHT was used for the other. Using the patented One-shot Kinetics approach, a dilution series of the analyte from 80 nM to 5 nM was injected for kinetic analysis. Following this the bound analyte was removed with phosphoric acid and the antibody surface was regenerated. Another injection of 80 nM analyte was applied in all six channels so that the analyte was uniformly reloaded on all 36 interaction spots. This was done to ensure a sufficient amount of analyte was bound to the sensor chip surface for subsequent MS analysis. Results are presented below (Figure 1, Table 1).
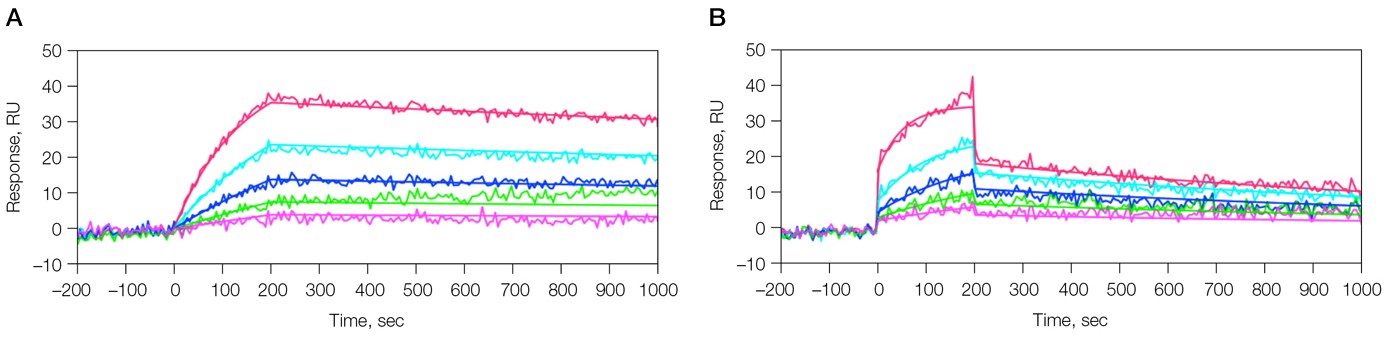
Fig. 1. Kinetic analysis of the interaction of 6E10 Ab and β-amyloid 1–40 fragment. Two sensorgrams were obtained under different analyte sample conditions. A, analyte in PBSHT; PBSHT running buffer; analyte concentrations of 80, 40, 20, 10, 5 nM; B, analyte in 10% human serum (diluted in the running buffer); PBSHT + 1% BSA running buffer; analyte concentrations of 80, 40, 20, 10, 5 nM.
Table 1. Kinetic constants for the binding of 6E10 Ab and β-amyloid 1-40 fragment.
|
Sample
|
ka (1/Ms) | kd (1/s) | KD (M) |
| β-amyloid 1–40 in PBSHT | 8.67 x 104 | 1.78 x 10-4 | 2.05 x 10-9 |
| β-amyloid 1–40 in 10% serum | 2.36 x 105 | 7.00 x 10-4 | 2.97 x 10-9 |
Biomolecule Identification Using MALDI-TOF-MS
The ProteOn sensor chips have a gold surface derivatized with a surface chemistry for biomolecule immobilization. The surface properties of the sensor chips make them excellent substrates for the analysis with a MALDI time-of-flight (MALDI-TOF) mass spectrometer (Figure 2).
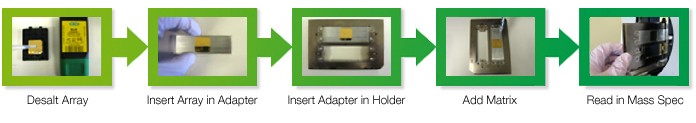
Fig. 2. The MALDI-TOF-MS workflow achieved using a single ProteOn sensor chip. The steps include desalting, insertion into the adapter and the holder, matrix addition, and MS analysis.
The MS analysis of β-amyloid 1–40 fragment on both sensor chips, including linear, reflectron, and MS/MS modes, were carried out with an UltrafleXtreme mass spectrometer (Bruker Corporation), leading to the successful identification of β-amyloid 1–40 fragment analyte (Figure 3).
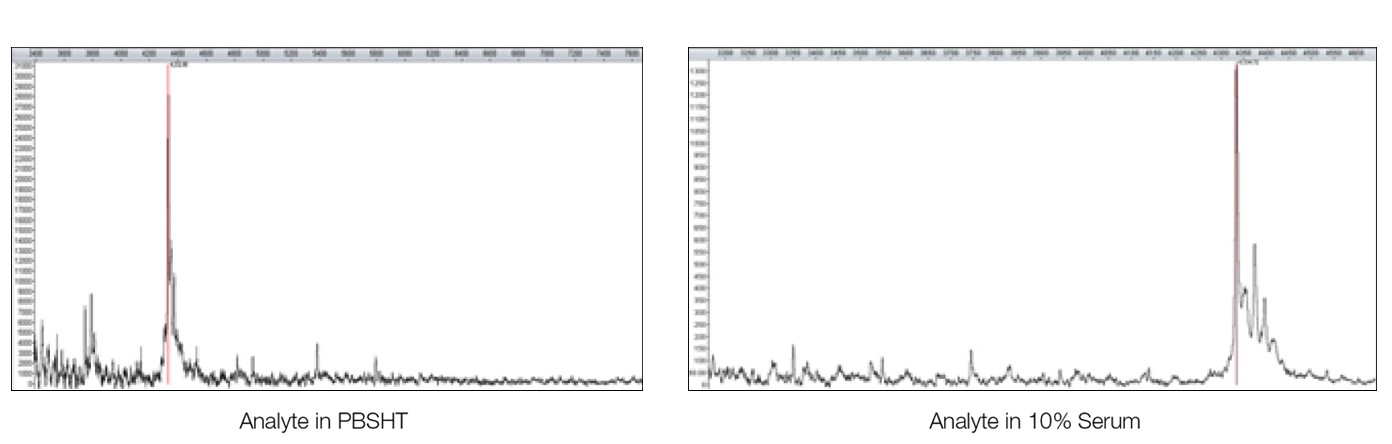
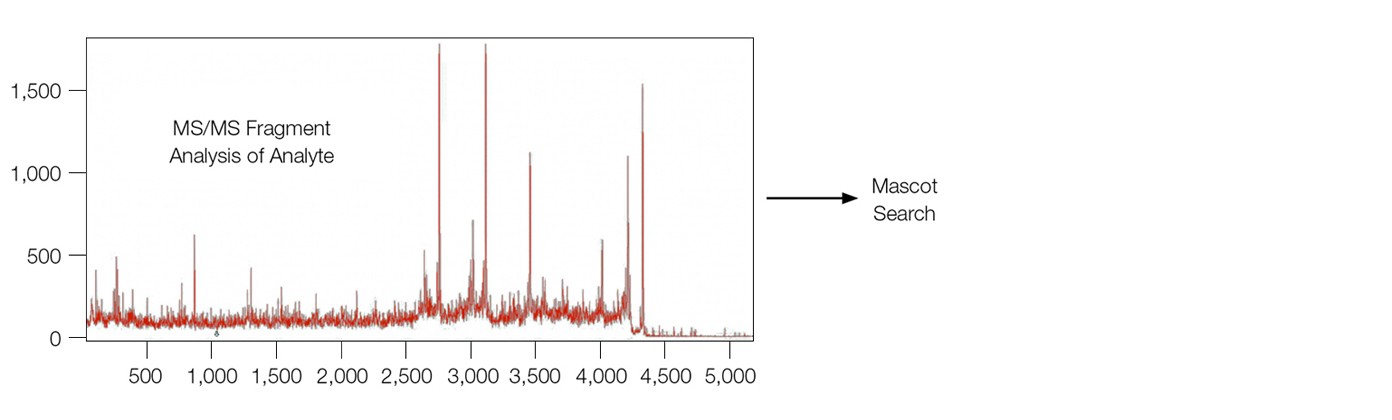
Fig. 3. MALDI-MS analysis of the analyte β-amyloid 1–40 fragment directly from the sensor chip surface. MS-MS fragment analysis lead to subsequent identification from a Mascot search. The sensor chip was desalted with water washes before adding the matrix (1 µl of 20% α-cyano-4-hydroxycinnamic acid in a 1:3 acetonitrile/1% trifluoroacetic acid solution).
Conclusion
This work proves the concept of direct MALDI-TOF-MS analysis from an SPR sensor chip surface without modification to the surface beyond desalting and matrix addition. The high quality experimental result shows the advantages of this novel SPR-MS method compared to traditional elution-based workflows in which downstream analysis by MS is problematic due to the small amounts of analyte bound to the sensor chip surface. Using this direct interrogation technique, we have shown a viable alternative approach to the SPR-MS analysis of protein samples.
UltrafleXtreme is a trademark of Bruker Corporation. Mascot is a trademark of Matrix Science Ltd. Tween is a trademark of ICI Americas Inc.

