Introduction
The data acquired using a surface plasmon resonance (SPR) sensor such as the ProteOn XPR36 protein interaction array system is processed using a multistep procedure. This article provides detailed guidelines for SPR data processing using the ProteOn system and the associated ProteOn Manager™ software. It describes sensorgram terms, sensorgram display, sensorgram processing, sensorgram referencing, and quality standards for processed sensorgrams. The steps of data processing and data analysis using ProteOn Manager software are outlined in the Appendix.
Interaction Sensorgram Terms
An SPR sensorgram is a graph of time-traced SPR responses during a biomolecular interaction analysis. The x-axis is time in seconds and the y-axis is SPR response in response units (RU). It should be noted that both ligand immobilization and analyte injection steps generate sensorgrams, but the sensorgram in the analyte injection step is more often used because it contains the ligand-analyte interaction information for affinity or kinetic analysis. Therefore, where not otherwise specified, “sensorgram” hereinafter refers to that generated in the analyte injection step.
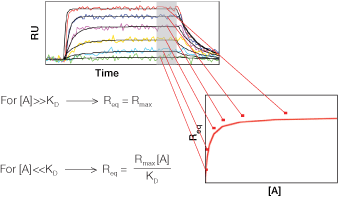
Fig. 1. A typical SPR sensorgram displaying the three sensorgram phases.
As illustrated in Figure 1, an SPR sensorgram can be divided into three different phases:
Baseline is the phase before the injection of the analyte. Running buffer flows over the sensor chip surface bearing the immobilized ligand and the baseline response is recorded.
Association is the phase during the injection of the analyte. The analyte flows over the sensor chip surface and binding occurs between the ligand and the analyte. A concave increasing response curve is produced. Depending on the binding kinetics of the interaction partners and the experimental conditions, the increasing response curve may or may not reach a plateau, which indicates that interaction equilibrium has been reached.
Dissociation is the phase after the injection of the analyte is finished. Running buffer flows over the sensor chip surface and washes off the analyte bound to the ligand. A convex decreasing response curve is produced. Depending on the binding kinetics of the interaction partners and the experimental conditions, the decreasing response curve may or may not return to baseline, which indicates complete dissociation of the analyte from the ligand surface.
Kinetic analysis requires the presence of all three phases, whereas equilibrium and concentration analyses do not. However, note that (1) the association phase must reach the interaction equilibrium plateau for an equilibrium analysis, and (2) the beginning of the association phase must be linear to reveal mass-transport limitations to kinetics in a concentration analysis.
Sensorgram Display
In the 6 × 6 experimental configuration of the ProteOn XPR36 system, 36 sensorgrams are produced simultaneously in each SPR experiment. In the sensorgram display, by default the sensorgrams are grouped by ligand in six sensorgram windows, or graphs. In the One-shot Kinetics™ approach, each graph contains a set of six dose-response sensorgrams for a dilution series of the analyte interacting with six identical ligand surfaces. During data processing, the grouping of sensorgrams can be adjusted in the Data Grouping screen, and individual sensorgrams can be removed from or added back to the sensorgram display in the Interaction screen.
The sensorgram graphs can be selected or deselected by clicking their blue title bars. In the menu bar, click View and Select All Graphs to select all the sensorgram graphs. While Auto data processing activities apply to all the sensorgram graphs, Selected data processing activities apply only to the selected sensorgram graphs.
Sensorgram Processing
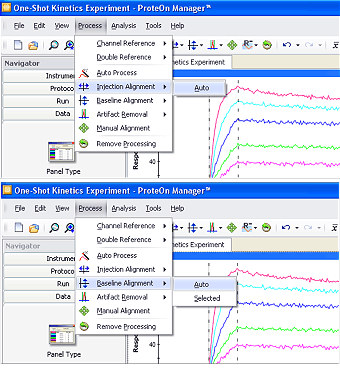
Fig. 2. The automatic injection alignment and baseline alignment functions offered in ProteOn Manager software.
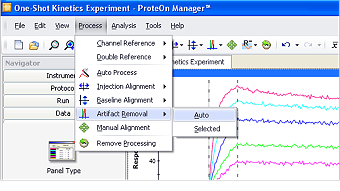
Fig. 3. The automatic artifact removal function offered in ProteOn Manager software.
1. Align the sensorgram set
To perform reliable kinetic analysis, a set of dose-response sensorgrams for the same interaction are typically analyzed together to minimize system deviations. This action requires the alignment of the graphs in a sensorgram set in both the vertical and horizontal dimensions. In scientific experiments, it is common to perform diagram or sensorgram alignment when processing multiple data groups in a dataset. The alignment action for SPR sensorgrams is similar to that performed for other bioanalytical technologies such as spectroscopy. ProteOn Manager software offers automatic injection alignment along the x-axis and baseline alignment along the y-axis, available in the Process menu, as shown in Figure 2.
Injection alignment adjusts all the sensorgrams to share the same starting point along the x-axis, thus removing time differences among the different sensorgrams. Unlike traditional serial-flow SPR systems, the ProteOn XPR36 system has a parallel flow design that allows for synchronized analyte injection across different flow channels. Therefore, injection alignment is typically achieved with very good accuracy.
Baseline alignment adjusts all the sensorgrams to the same zero-baseline level along the y-axis, thus removing slight baseline-level differences among the sensorgrams resulting from previous steps. Although the automatic baseline alignment function processes entire sensorgrams by default, it is possible to align the sensorgrams based on the values in a selected region. To define the selected region, click and drag in the sensorgram graph. Click Process, select Baseline Alignment, and choose Selected in the submenu to perform baseline alignment for the selected region.
In addition to the alignment options described above, manual correction may be applied to fine-tune the sensorgrams. To perform the manual sensorgram alignment, click Process and select Manual Alignment. Individual sensorgrams can then be moved when selected with the mouse.
2. Remove the artifacts
Sensorgram artifacts, usually spikes caused by tiny air bubbles in the analyte solution, are sometimes present and should be removed. Note that artifact here refers to a response deviation over a very small time period. If a significant portion of the sensorgram deviates from the expected response, the trial should be re-run.
ProteOn Manager software offers automatic artifact removal that flattens these artifacts to restore sensorgram integrity, as illustrated in Figure 3. Although the automatic artifact removal processes entire sensorgrams by default, it is possible to process a selected region of the sensorgrams. To define the selected region, click and drag in the sensorgram graph. Click Process, select Artifact Removal, and choose Selected in the submenu to perform artifact removal within the selected region.
For ease of use, ProteOn Manager software allows the user to conduct all the sensorgram processing steps with a single command. Selecting Auto Process in the Process menu sequentially performs injection alignment, baseline alignment, and artifact removal.
Sensorgram Referencing
Sensorgram referencing is the most important step in data processing. The subtraction of references removes artifacts of refractive index change (bulk effect) from the analyte sample, nonspecific binding (NSB) of the analyte and impurities on the sensor chip surface, and changes of the ligand surface. There are two types of referencing in SPR analysis: blank surface referencing and blank buffer referencing. A blank surface reference is used to correct for bulk effect and NSB, and a blank buffer reference is used to correct for baseline drift resulting from the changes of the ligand surface.
The novel 6 × 6 experimental configuration in the ProteOn XPR36 system offers a comprehensive set of referencing options. The referencing options can be selected from the Process menu, as shown in Figure 4. The available referencing options are listed below.
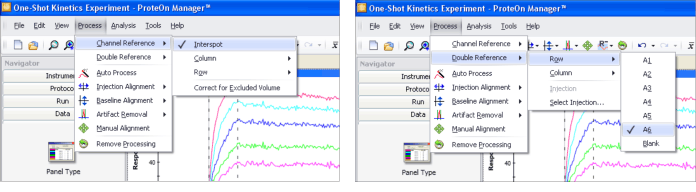
Fig. 4. ProteOn Manager software presents all the referencing options available in the ProteOn XPR36 system for selection.
1. Blank surface referencing (also known as channel referencing)
Blank surface referencing is performed on a blank surface (either an empty or an irrelevant protein-coated surface) with an analyte solution flowing over it. The reference responses are collected on blank surfaces during the analyte injection (that is, blank surface reference = blank surface + analyte solution), as shown in Figure 5. Blank surface referencing is used to correct for bulk effect and NSB.
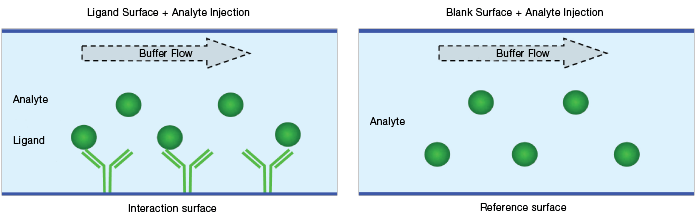
Fig. 5. Schematic diagram of a blank surface reference.
The ProteOn XPR36 system offers two blank surface referencing options, as illustrated in Figure 6.
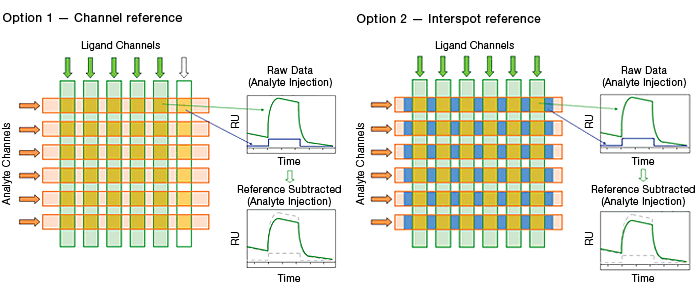
Fig. 6. The two blank surface referencing options in the ProteOn XPR36 system.
Channel referencing is the reference method traditionally used in commercial SPR biosensors. It involves reserving a portion of the potential interaction surfaces for use as blank surfaces.
Interspot referencing is unique to the ProteOn XPR36 system. Instead of consuming potential interaction surfaces, this reference method employs the interval surfaces adjacent to interaction surfaces. Compared with the traditional channel reference, the interspot reference has the advantages of immediate proximity to interaction spots and the conservation of interaction spots. The immediate proximity enhances the referencing quality.
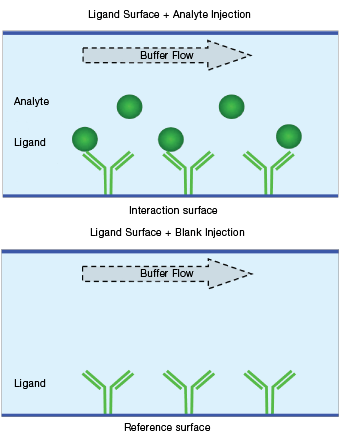
Fig. 7. Schematic diagram of a blank buffer reference.
2. Blank buffer referencing (also known as double referencing)
Blank buffer referencing is performed on a ligand surface with a blank buffer (either a running buffer or a negative control sample) flowing over it. The reference responses are collected on ligand surfaces during a blank buffer injection (that is, blank buffer reference = ligand surface + blank buffer), as shown in Figure 7. Blank buffer referencing is used to correct for baseline drift resulting from the changes of the ligand surface.
The ProteOn XPR36 system offers two blank buffer referencing options, as illustrated in Figure 8.
Injection referencing is the reference method traditionally used in commercial SPR biosensors. It requires a blank buffer injection performed prior to the analyte injection.
Real-time double referencing is unique to the ProteOn XPR36 system. This method employs a blank buffer injection in parallel with the analyte injection. Compared with the traditional injection reference, the real-time double reference has the advantages of accurate monitoring of possible changes on ligand surfaces and saving time by eliminating the additional blank buffer injection. The accurate monitoring of ligand surfaces greatly enhances the referencing quality, especially in the cases of capture surfaces, where reversible capture of the ligand is employed and exponential baseline decay is often observed.
Referencing options should be selected in the experiment design phase, as the reference surfaces are created in ligand immobilization and analyte injection steps. A combination of blank surface and blank buffer referencing is usually applied to yield high-quality SPR results. This combination is implemented by sequentially subtracting one blank surface reference and one blank buffer reference. The ProteOn XPR36 system offers the flexibility of selecting any combination of the four references to optimize data processing.
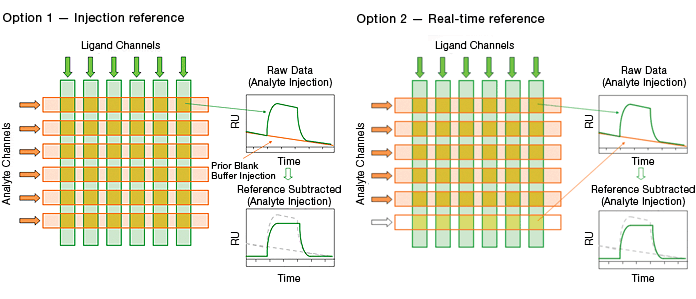
Fig. 8. The two blank buffer referencing options in the ProteOn XPR36 system.
3. Excluded volume correction
Excluded volume correction (EVC) is not an independent referencing option but rather a calibration with a blank surface reference. This calibration is applied when a cosolvent with a high refractive index, such as DMSO, is used in an analyte solution to increase the analyte solubility. A high refractive index cosolvent may produce a larger bulk effect on a reference surface than on an interaction surface due to the volume exclusion of the cosolvent by the ligand on the interaction surface. This inconsistency can be resolved by the EVC calibration. Please refer to Bio-Rad bulletin 5822 for a detailed explanation of this calibration and experimental guidance.
Quality Standards for Processed Sensorgrams
The following standards are used to judge the quality of processed sensorgrams:
- Processed sensorgrams — the sensorgrams are aligned in both dimensions, and artifacts, such as air bubbles, are removed. This processing requires both good-quality raw sensorgrams and appropriate software functions.
- Good choice of referencing — both blank surface referencing (channel referencing) and blank buffer referencing (double referencing) are appropriately performed. The experimental design must ensure the incorporation of the correct referencing options. The referenced sensorgrams should not show bulk effects or baseline drift. Although it is not required, the best practice is to have no response jump present between the end of association and the beginning of dissociation phases.
- Sufficient interaction time — the interaction time or the time of analyte injection in the association phase is long enough to show curvature, and the running buffer injection time in the dissociation phase is long enough to show adequate response decrease to resolve the dissociation rate constant. The choice of appropriate injection conditions, including interaction time, analyte concentration, and injection flow rate, is based on the user’s understanding of the interaction. For example, the user can determine the binding affinity and ligand-analyte complex stability by obtaining this information from either preliminary experimental trials or literature values. This consideration is essential for accurate sensorgram fitting.
Note: The steps of data processing and data analysis using ProteOn Manager software are outlined in the Appendix.
The ProteOn XPR36 protein interaction array system is covered by Bio-Rad patents, including United States patent numbers 8,111,400, 8,105,845, 7,999,942, and 7,443,507.
This product or portions thereof is manufactured and sold under license from GE Healthcare under United States patent numbers 5,492,840, 5,554,541, 5,965,456, 7,736,587, and 8,021,626, and any international patents and patent applications claiming priority.

