Introduction
Ligand-capture approaches are often utilized in biomolecular interaction analysis using surface plasmon resonance (SPR) biosensors. In many cases, biotin is used as an affinity tag for general ligand-capture approaches because of its various advantages. However, a drawback to using biotin is that it makes surface regeneration of sensor chips difficult. In this article, we propose a novel method to enable a surface regeneration capability for the biotinylated ligand-capture approach. It employs a two-layer capture strategy: first using nickel (II)-activated tris-nitriloacetate (tris-NTA) to capture histidine-tagged streptavidin, and then using streptavidin to capture a biotinylated ligand protein. An experiment was conducted to verify the performance of this novel biotinylated ligand-capture method and the results are presented here.
Surface plasmon resonance (SPR) biosensors measure biomolecular interactions in a label-free and real-time manner. A typical SPR experiment involves first immobilizing a ligand (a biomolecule such as a protein or a nucleic acid) to a functionalized sensor chip and then flowing an analyte (another biomolecule such as a protein or a small molecule) over the chip surface to investigate the binding affinity and kinetics of the analyte to the ligand. Binding of the analyte to the ligand is tracked by following changes in the SPR signal over time using time-traced graphs called sensorgrams. Fitting the sensorgram to a suitable kinetic binding model allows the calculation of kinetic parameters such as the association rate constant (ka) and dissociation rate constant (kd), as well as the dissociation equilibrium constant (KD).
Ligand capture refers to the noncovalent capture of a ligand using a capture reagent and is often employed in SPR experiments. The capture reagent is typically covalently linked to the chip surface. Thus, the ligand is immobilized to the chip surface through reversible binding to the capture reagent. This method can provide high ligand activity on the chip surface because a suitable capture mechanism can be selected to facilitate good orientation of the ligand molecules. In addition, it offers the possibility of regenerating the chip surface by completely removing the reversibly bound ligand for the re-use of the sensor chip.
It is common to use an antibody as the capture reagent to immobilize the ligand (Homola 2008). However, this type of ligand capture requires a specific antibody for each ligand. Thus, to facilitate general ligand capture, affinity tags are often employed. An affinity tag is a domain attached to the ligand protein that facilitates its binding to the capture reagent. In biosensor assays, typical affinity tags include biotin, histidine-tag (polyhistidines), etc. (Morgan and Taylor 1992; Sigal et al. 1996, and Wegner 2003). Biotin is the preferred affinity tag in many cases due to an available collection of mature methods for biotinylation. In addition, its high affinity for avidin family proteins, such as avidin, streptavidin, and NeutrAvidin, allows stable binding to a capture reagent of that family (Leckband 2000). (Streptavidin was used in the current experiment and is used throughout this article to represent avidin family proteins.) When biotin is used as an affinity tag in an SPR experiment, streptavidin is first covalently linked to the chip surface as the capture reagent, and then a biotinylated ligand is captured to establish a ligand surface for the interaction analysis that follows.
The high affinity between biotin and streptavidin, characterized by a very low dissociation equilibrium constant (KD) at the level of 10–14 M, minimizes the baseline drift caused by ligand leaching from the chip surface (Chilkoti and Stayton 1995). However, it also results in difficult surface regeneration of sensor chips because it is impossible to remove the biotinylated ligand from streptavidin under normal conditions. Thus, applying the biotinylated ligand-capture approach in an SPR experiment is costly because sensor chips can be used only once.
An available solution to enable surface regeneration is to use a secondary capture mechanism to noncovalently capture streptavidin to the chip surface. In this case, there are two capture reagents involved: streptavidin as the primary reagent to capture the biotinylated ligand and a secondary reagent to capture streptavidin. The secondary capture reagent should be covalently linked to the chip surface to guarantee stability of the two-layer capture. As long as the binding mechanism between the secondary capture reagent and streptavidin is reversible, the biotinylated ligand together with streptavidin can be completely removed from the chip surface, enabling surface regeneration. The concept is illustrated in Figure 1A.
In order to implement this two-layer capture strategy, a good capture mechanism between the secondary capture reagent and streptavidin is of the utmost importance. It must provide enough binding stability to minimize baseline drift due to ligand leaching and allow an easy way to reverse the capture for surface regeneration. The CAPture kit offered by GE Biacore employs DNA hybridization as the secondary capture mechanism to immobilize an avidin family protein (Papalia and Myszka 2010). The surface regeneration in this case is realized by triggering DNA dehybridization. Although this approach allows the re-use of sensor chips, it does not work in a low ionic strength environment because it will cause DNA dehybridization that is detrimental to the capture of the biotinylated ligand. In addition, it does not significantly reduce the cost of an SPR experiment due to the additional expense of purchasing the special reagents.
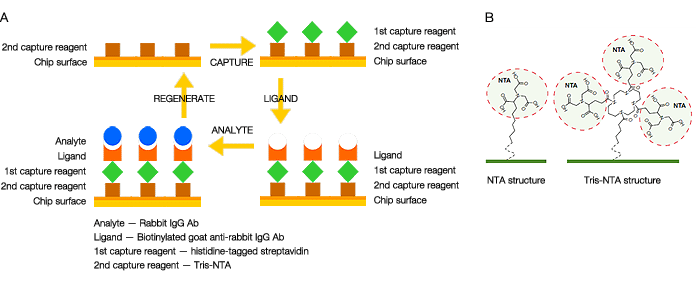
Fig. 1. A, the two-layer capture strategy that allows surface regeneration of sensor chips for biotinylated ligand capture; B, NTA and tris-NTA chemical structures bound to SPR sensor chips. The ProteOn HTG and HTE chips have tris-NTA covalently linked to the chip surface.
In this article, we propose a new method to realize the two-layer capture strategy. This new method employs histidine-tagged streptavidin as the primary and nickel (II)-activated nitrilotriacetate (NTA) as the secondary capture reagent. Because the binding between a histidine-tag and NTA can be easily reversed by removing the nickel (II) ions with EDTA, surface regeneration of the sensor chips is facilitated with this method. However, it is known that the binding affinity between a histidine tag and NTA is typically not high. To overcome this drawback, the novel tris-NTA (3 x NTA) surface chemistry is employed. This surface chemistry facilitates the formation of a nickel (II)-activated tris-NTA complex that intercalates at three points with a single histidine tag to significantly enhance binding affinity (Lata and Piehler 2005 and Lata et al. 2006). A comparison of the NTA and tris-NTA chemical structures is shown in Figure 1B. The dissociation rate constant of a captured histidine-tagged ligand protein is typically reduced to the level of 10-5 s-1, while the surface regeneration capability is maintained. The two-layer capture strategy employing histidine-tagged streptavidin can be implemented using the tris-NTA surface chemistry. Baseline drift due to ligand leaching in this case is trivial.
Experiment and Results
In this work, the SPR biosensor ProteOn™ XPR36 protein interaction array system was used as the experimental platform. The ProteOn XPR36 system provides a novel 6 x 6 interaction array that analyzes 36 biomolecular interactions simultaneously, using the One-shot Kinetics™ technology. It enables efficient experimental optimization, rapid data acquisition, and multiple referencing options for high-quality experimental results (Bravman et al. 2006). The ProteOn HTG and HTE chips were used to test the biotinylated ligand-capture approach. These sensor chips feature the tris-NTA surface chemistry. The tris-NTA complexes are covalently linked to the chip surface (Rabkin et al. 2012 and Bronner et al. 2012).
An interaction analysis was tested for the proof of principle of this new method. The experiment included four sequential steps: (1) activation of the secondary capture reagent, tris-NTA, by nickel sulfate; (2) capture of the primary capture reagent, histidine-tagged streptavidin (Abcam ab78833), to the chip surface; (3) capture of the ligand, biotinylated goat anti-rabbit IgG (Fc fragment specific) antibody (Jackson ImmunoResearch 111-065-046), to streptavidin; (4) injection of the analyte, rabbit IgG (Sigma-Aldrich, St. Louis, MO), at five concentrations for the interaction analysis. After the interaction analysis was completed, surface regeneration was implemented following instructions from the manufacturer’s user guide. The original chip surface with only tris-NTA was restored and the sensor chip could be used for a repeated experimental cycle. The experimental results of three repeated cycles using an HTG sensor chip and the kinetic and equilibrium constants obtained from sensorgram fitting are shown in Figure 2. In each experimental cycle, three independent ligand channels (L1, L2, and L3) carrying out the same interaction analysis are presented. Both channel-to-channel reproducibility and cycle-to-cycle reproducibility are verified by the consistent experimental results.
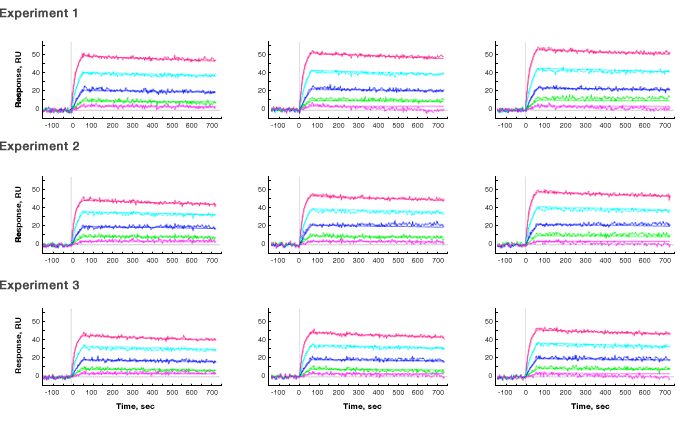
Fig. 2. Experimental results of the new two-layer capture strategy. Three repeated experimental cycles were performed and the sensorgrams with fitting lines in the analyte injection steps are shown. In each experimental cycle, three ligand channels carrying out the same interaction analysis are presented. The sensor chip is an HTG chip, the primary capture reagent is tris-NTA immobilized on the chip surface, the secondary capture reagent is histidine-tagged streptavidin, the ligand is biotinylated goat anti-rabbit IgG antibody, and the analyte is rabbit IgG antibody. The secondary capture reagent was captured to the response level of ~230 RU, the ligand was captured to the response level of ~160 RU, and the analyte concentrations were 30 nM (—), 12 nM (—), 4.8 nM (—), 1.9 nM (—), and 0.8 nM (—). The running buffer was PBS with 0.05% Tween 20.
The kinetic and equilibrium constants obtained from sensorgram fitting are shown in the table.
| Parameter | ka Ms-1 | kd s-1 | KD M | |
| Channel L1 | ||||
| Experiment 1 | 1.39 x 106 | 1.47 x 10-4 | 1.06 x 10-10 | |
| Experiment 2 | 1.70 x 106 | 1.38 x 10-4 | 8.12 x 10-11 | |
| Experiment 3 | 1.68 x 106 | 1.67 x 10-4 | 9.92 x 10-11 | |
| Channel L2 | ||||
| Experiment 1 | 1.40 x 106 | 1.49 x 10-4 | 1.06 x 10-10 | |
| Experiment 2 | 1.63 x 106 | 1.38 x 10-4 | 8.47 x 10-11 | |
| Experiment 3 | 1.57 x 106 | 1.64 x 10-4 | 1.04 x 10-10 | |
| Channel L3 | ||||
| Experiment 1 | 1.37 x 106 | 1.23 x 10-4 | 8.96 x 10-11 | |
| Experiment 2 | 1.54 x 106 | 1.20 x 10-4 | 7.80 x 10-11 | |
| Experiment 3 | 1.55 x 106 | 1.52 x 10-4 | 9.83 x 10-11 | |
| Overall Average | 1.54 ± 0.13 x 106 | 1.44 ± 0.16 x 10-4 | 9.41 ± 1.09 x 10-11 | |
| Overall CV* | 8% | 11% | 12% | |
* Calculated for all nine results — three injections on three ligand channels.
The same interaction analysis was also tested on an HTE chip and similar results were obtained. In order to verify the accuracy of the kinetic analysis on the HTG and HTE chips, the same interaction analysis was performed on a different sensor chip, the ProteOn NLC chip. The NLC chip contains NeutrAvidin, so the biotinylated ligand could be directly captured to the chip surface without using histidine-tagged streptavidin. The average kinetic and equilibrium constants in three repeated experimental cycles are: ka 1.34 ± 0.10 x 106 M-1s-1, kd 1.49 ± 0.27 x 10-4 s-1, KD 1.12 ± 0.28 x 10-10 M-1. The results are very close to those obtained using the two-layer capture strategy on an HTG chip shown in the table above.
The SPR experiment reported in this article has proved that the new two-layer capture strategy utilizing histidine-tagged streptavidin and tris-NTA sensor chips works effectively for biomolecular interaction analyses. This method allows for the surface regeneration of sensor chips by the removal and rebuilding of the biotinylated ligand-capture surface chemistry. Reproducible experimental results have verified the accuracy of interaction analysis and the capability for surface regeneration. We believe that more experiments analyzing biomolecular interactions can be achieved using this method.
References
Homola J (2008). Surface plasmon resonance sensors for detection of chemical and biological species. Chem Rev 108, 462–493.
Morgan H and Taylor DM (1992). A surface plasmon resonance immunosensor based on the streptavidin-biotin complex. Biosens Bioelectron 7, 405–410.
Sigal GB et al. (1996). A self-assembled monolayer for the binding and study of histidine-tagged proteins by surface plasmon resonance. Anal Chem 68, 490–497.
Wegner GJ et al. (2003). Fabrication of histidine-tagged fusion protein arrays for surface plasmon resonance imaging studies of protein-protein and protein-DNA interactions. Anal Chem 75, 4740–4746.
Leckband D (2000). Measuring the forces that control protein interactions. Ann Rev Biophys Biomol Struct 29, 1–26.
Chilkoti A and Stayton PS (1995). Molecular origins of the slow streptavidin-biotin dissociation kinetics. J Am Chem Soc 117, 10622–10628.
Papalia G and Myszka D (2010). Exploring minimal biotinylation conditions for biosensor analysis using capture chips. Anal Biochem 403, 30–35.
Lata S and Piehler J (2005). Stable and functional immobilization of histidine-tagged proteins via multivalent chelator headgroups on a molecular poly(ethylene glycol) brush. Anal Chem 77, 1096–1105.
Lata S et al. (2006). Monitoring the dynamics of ligand-receptor complexes on model membranes. J Am Chem Soc 128, 6–7.
Bravman T et al. (2006). Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal Biochem 358, 281–288.
Rabkin et al. (2012). The ProteOn HTG sensor chip: novel surface for stable capture of histidine-tagged proteins for protein-protein interaction analysis. Bio-Rad Bulletin 6132.
Bronner et al. (2012). The ProteOn HTE sensor chip: novel surface for stable capture of histidine-tagged proteins for protein-small molecule interaction analysis. Bio-Rad Bulletin 6254.
Biacore is a trademark of GE Healthcare group companies. NeutrAvidin is a trademark of Thermo Fisher Scientific, Inc.

