For most applications, purifying the protein of interest has become routine. Proteins can be overexpressed, affinity tagged, and purified using commercially available reagents and vendor-provided protocols. Specialized expression systems promote disulfide bond formation, allow for posttranslational modifications, and minimize protein degradation. Tags can be added to increase stability and solubility, while gravity columns have been replaced with user-friendly chromatography systems preprogrammed with method templates.
But for some applications, protein purification continues to be challenging. Purifying proteins for structural studies as well as integral membrane proteins and untagged proteins from their native sources remains a daunting task. Here we discuss general guidelines, resin properties, and advances in chromatography automation that can greatly simplify the purification of even these most-difficult-to-purify proteins.
Obtaining high yields and purity for structural studies
NMR and crystallography are powerful tools for determining protein structures, but both techniques require high protein concentrations (in the 5–50 mg/ml range) and purity (>95% pure by SDS-PAGE). These studies also demand extraordinary reproducibility between protein preparations. Tiny differences in purity — or even in conformational state — can affect crystal growth, and differences in sample composition are financially draining when NMR studies need to be repeated, as this technique requires isotopic labels that are very expensive. Structural genomists face an additional obstacle: the need to obtain high protein concentration and purity at incredibly high throughput levels. Protocols created as part of the NIH Protein Structure Initiative pilot phase, for example, were developed to set up 2,880 different crystallization experiments per hour.
Effectively exploring purification space
To identify purification conditions that yield the required protein concentration and purity often requires screening many resins and buffer systems. Figure 1 provides an example of a typical purification decision tree. The ability to explore not only different chromatography modes but also different bead sizes can lead to higher purity. However, exhaustive exploration of purification options is nearly impossible using traditional chromatography systems that allow the operator to set up and program only a single chromatography run at a time.
This problem has been addressed with automation-friendly systems like the NGC™ Chromatography System. The accompanying ChromLab™ Software’s scouting feature enables quick and easy generation of pH, column, and %B scouting runs, resulting in faster optimization, more exhaustive exploration of purification space, and thus maximized protein yields and purity. To further increase yields and purity, multiple variables can be scouted simultaneously using design of experiment (DoE). Such approaches allow identification of positive and negative correlations between changes in different purification parameters and generation of mathematical models that can predict purification outcomes for nontested conditions.
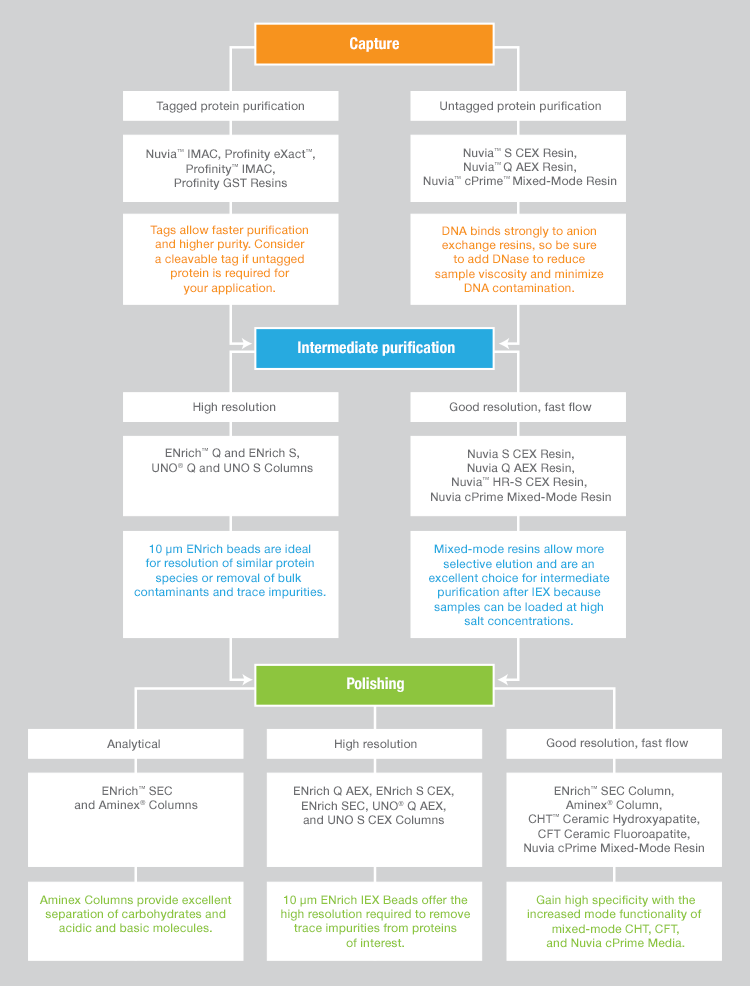
Fig.1. Resin type and size decision tree.
Reducing batch-to-batch variability
Once optimal purification conditions have been determined, they need to be implemented in a reproducible manner. At this stage researchers often find that even when using the exact same methods, sample purity or concentration varies. Some applications tolerate these subtle differences in purification output, but structural studies are not so forgiving. It is not uncommon for a new protein preparation to no longer yield crystals.
A main source of this variability is inconsistencies introduced when manually pooling fractions. The solution lies in limiting manual intervention by automating multicolumn purification. The NGC System’s multidimensional (Multi-D) chromatography capability, for example, removes the need to analyze and pool fractions manually by allowing incorporation of multiple column steps into a single automated method with independent pressure monitoring and reverse elution capability. Using a single sample inlet valve, up to seven different samples can be purified with the push of a button; by adding a second sample inlet valve this number can be increased to 14, and with the NGC™ Autosampler up to 96 samples can be run in an automated fashion. Advances in automation, such as Multi-D, thus allow scientists to not only limit batch-to-batch variability but also increase purification throughput.
See how automating multicolumn purifications can improve protein purification.
Optimizing expression levels and solubility of membrane proteins
Membrane proteins are another class of notoriously difficult-to-purify proteins. Even though they make up 20–30% of eukaryotic proteomes and ~50% of drug targets, they account for only about 1% of proteins in the Protein Data Bank.
Expressing integral membrane proteins in E.coli or other expression systems can be challenging as it often results in aggregation, misfolding, improper membrane insertion, toxicity, or a combination of the above. In addition, the hydrophobic surfaces that allow these proteins to be inserted into membranes can prevent them from being soluble in aqueous solutions. For each membrane protein, conditions have to be found that release it from the membrane while maintaining its structure and activity. A key part of successfully purifying membrane proteins is thus optimizing both expression and solubilization conditions.
Efficiently assessing expression hosts and constructs
To assess expression conditions, C-terminal affinity tags can be used to enrich for the often low-expressing protein of interest; N-terminal tags should be avoided as they can interfere with membrane insertion. Different expression strains and constructs should be tested in parallel to identify conditions that yield the highest amount of properly inserted membrane protein. When designing constructs consider varying plasmid copy number and promoter strength, and when possible test homologs from different species to identify the best-expressing construct. This can quickly yield large numbers of samples that need to be tested. Small-scale 96-well culture formats and small-volume spin columns or prepacked 96-well chromatography filter plates allow for fast and economical screening of many expression conditions.
Accelerating solubility screening
In addition to testing different strains and constructs, solubilization conditions need to be optimized once favorable expression conditions are determined. An autosampler coupled to an automation-friendly chromatography system, such as the NGC Autosampler coupled to the NGC Chromatography System, can accelerate this process as samples subjected to different detergent conditions can be assembled in the autosampler and run sequentially in an automated fashion. Aggregation state can be assessed using dynamic light scattering, size exclusion chromatography, or a combination of the two techniques. The NGC System simplifies analysis by dynamic light scattering as it allows you to export data collected on the NGC UV Detector to third-party light-scattering analysis software. When possible, resins with hydrophilic base bead characteristics, such as Nuvia, ENrich, and UNOsphere™ Resins, should be used to reduce nonspecific binding of membrane proteins to the resin.
By using tools such as spin columns, autosamplers, and automation-friendly chromatography systems, expression and solubilization conditions can be explored more thoroughly, increasing the chances of identifying conditions that allow purification of workable amounts of soluble and active protein.
Developing purification strategies for untagged native-source proteins
Sometimes it is not possible to overexpress tagged proteins in heterologous expression systems. These cases require purification of untagged proteins that are constitutively expressed and thus usually present only at low concentrations. Here efficient column and buffer scouting is a must.
Optimizing purification conditions through resin, pH, and %B scouting
A common purification strategy for untagged proteins is ion exchange (IEX) for capture, followed by hydrophobic interaction chromatography (HIC) for intermediate purification and size exclusion chromatography (SEC) as a final polishing step. Mixed-mode resins, such as CHT Ceramic Hydroyxapatite and Nuvia cPrime Resin, may yield better results than IEX or HIC as they allow greater control of elution and separation of contaminants. Because mixed-mode resins interact with proteins through several modes instead of just one, even difficult product contaminants such as charge variants can be separated from one another. Selectivity of mixed-mode resins can be further modulated by additives that preferentially affect the interaction of proteins of interest and contaminants with different resin functional groups. Mixed-mode resins are particularly advantageous as an intermediate step because the high-salt ion exchange eluate can be loaded directly onto the mixed-mode column without buffer exchange or dilution.
The protein of interest pI and lysis buffer pH will determine whether anion or cation exchange should be used as a capture step. Many soluble proteins have a pI of ~8.5 in E. coli, making cation exchange (CEX) an excellent choice for capture, especially since CEX allows greater separation of proteins of interest from negatively charged nucleic acids. It is advisable to test IEX resins with varying bead sizes; smaller beads will allow greater separation while larger beads permit faster flow rates. Most vendors will provide IEX resins with varying base bead sizes (Table 1).
For an initial capture step larger bead sizes are generally required because they accommodate viscous crude lysates and a fast flow rate. But keep in mind that larger beads also have lower binding capacity, which can prove problematic for recovery. Adding DNase to your lysate will decrease sample viscosity and allow faster flow rates and/or the use of smaller beads. It is also advisable to test different base bead chemistries as proteins can interact with resins not only through functional groups but also with the base bead itself.
Table 1. Base bead characteristics of Bio-Rad chromatography resins.*
| Product | Derivatives | Available Base Bead Sizes, µm |
| CHT Ceramic Hydroxyapatite | Hydroxyapatite, Type I and Type II | 20, 40, 80 |
| MPC™ Ceramic Hydroxyfluoroapatite | Partially fluorinated, Type I | 40 |
| CFT Ceramic Fluoroapatite | Fluorinated, Type II | 40 |
| Nuvia Resin | AEX, CEX, mixed-mode, IMAC | 50, 70, 85 |
| ENrich Resin | AEX, CEX, SEC | 10, 11 |
| UNO® Matrix | AEX, CEX | monolith |
* Most of these resins are available in prepacked columns and/or cartridges.
Once an optimal capture IEX resin has been identified, it is advisable to screen binding buffer pH as it affects the ionization state of proteins and thus their ability to interact with IEX resins. Binding pH screening should be followed by %B screening to optimize elution conditions. Chromatography systems equipped with buffer blending valves and scouting features, such as the NGC System, allow fast and convenient screening of many different buffer conditions and thus more exhaustive exploration of purification options. Similar optimization should be performed for the intermediate and final polishing steps.
In some cases, when sample amounts are limited, small spin columns or 96-well filter plates prepacked with chromatography resins may be preferable for buffer and pH screening, as these allow screening using smaller sample volumes than those required by automated chromatography systems.
Saving time by planning ahead
Whether you are trying to purify membrane proteins, untagged proteins, or proteins for structural studies, a key to purification success is to plan ahead. Map out potential purification strategies to avoid wasting time and resources by generating constructs that will be of no use for your protein of interest. Research and take advantage of the wide variety of commercially available cloning tools, expression systems, and chromatography resins as well as screening tools and automation capabilities. Using the right tools for your particular purification challenge can greatly increase the chances of successfully purifying even the most challenging targets.
For detailed information about how Bio-Rad can help, including information about our chromatography columns, systems, and resins as well as webinars, application notes, and instructional videos, visit our Chromatography Resource Page.

