Introduction
The ProteOn XPR36 protein interaction array system was introduced in 2006 to provide novel array technology to researchers conducting label-free analysis of proteins. The ProteOn system is a high-throughput surface plasmon resonance–based screening tool for a wide range of applications from small-molecule drug discovery to antibody kinetic ranking, epitope binning, and epitope mapping. The ProteOn system’s ability to process multiple samples in parallel allows users to generate kinetic data faster than traditional serial-processing SPR platforms, so researchers are able to efficiently screen, characterize, and optimize 36 interactions at once. Since its release, ProteOn system customers have used the system to accelerate a variety of research efforts, including kinetic evaluation of cancer therapeutic antibodies (Nahshol et al 2008), understanding the development of Alzheimer’s disease (Di Fede et al 2009), and characterizing prion proteins from mouse brain captured on the chip by specific antibodies (Biasini et al 2009 and Balducci et al 2010). The evolution of this product platform has been greatly influenced by product users, who have continually applied the technology in new ways. Dr Yasmina Abdiche’s exploration of a method to go above and beyond the conventional 6 x 6 analysis format of the system and develop a method that allows 36 unique interactions to be studied simultaneously on one array is such an example.
Uncovering the Potential for 36 x 36 Interactions
Abdiche graduated from Oxford University with a PhD in biological chemistry, and completed post-doctoral work in the laboratory of Dr David Myszka at the University of Utah, where she focused on developing protocols to analyze small-molecule interactions with proteins and lipids using label-free biosensors. During that time she was involved in establishing carbonic anhydrase as a model system in the biosensor community. She joined Rinat Neuroscience in 2004, a small biotech company that was acquired by Pfizer, Inc. in 2006, to lead the biosensor group within the company’s protein engineering department. This group uses various label-free technologies including the ProteOn system, to provide core support to multiple projects at all stages of research. One of the primary objectives of her group is to kinetically screen and epitope bin antibodies to aid in Pfizer’s drug discovery efforts. “Each instrument has its own strengths and weaknesses, and we are lucky to have a repertoire of label-free technologies to choose from in deciding which platform is most appropriate for a given application,” Abdiche explains.
Although Abdiche has appreciated the high-throughput capacity of the ProteOn system, her responsibilities to fulfill the needs of different groups working on multiple projects have driven her to continue to improve efficiency without sacrificing quality in her core workflow. “The drug discovery environment is increasingly competitive,” she says, “with significant pressure to generate reliable data as rapidly as possible to answer fundamental questions about the antibodies that we work with, because being first-to-market is critical.” Abdiche harnessed the versatility of the ProteOn system to enable a significantly higher throughput than what could be achieved through its traditional use. What she devised was a method to immobilize 36 independent ligands on each array to exploit its full ligand capacity (Figure 1). This method is described in full in the “Creating ligand arrays on the ProteOn for kinetics” section of Abdiche et al. 2011.
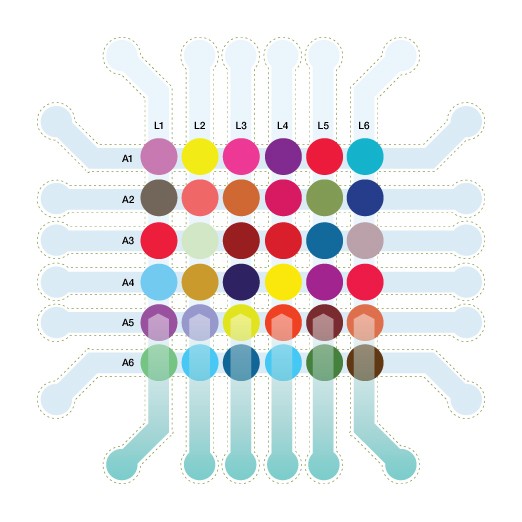
Fig. 1. Simultaneous interaction analysis on 36 unique ligands.
Validating the Methodology
When theorizing a way to use the ProteOn system to analyze interactions on 36 independent ligands simultaneously, Abdiche’s first challenge was conducting proof-of-concept experiments to validate her hypothesis. She designed experimental conditions for immobilizing two unrelated sets of antibodies (Groups I and II) that targeted different antigens (analytes 1 and 2). The antibodies chosen for this validation process had been well-characterized by her group and regeneration was needed only for the interactions of analyte 2 with the Group II antibodies.
Abdiche set up four different validation studies to assess the reproducibility and quality of the binding kinetics that could be generated using the 36-ligand array approach on the ProteOn system: 1) spot-to-spot reproducibility by immobilizing the same ligand onto different coordinates within the array, 2) comparison of different injection methodologies (short-and-long or multi-cycle analyses), 3) analyte reproducibility, and 4) equilibrium binding analysis.
Spot-to-Spot Reproducibility Studies
Interaction analysis on the Group I array was highly reproducible cycle-to-cycle on a per spot basis (Figure 2A). The data from both Group I and Group II antibody arrays showed excellent spot-to-spot reproducibility for any given antibody that was immobilized on two different coordinates within an array (Figure 2).
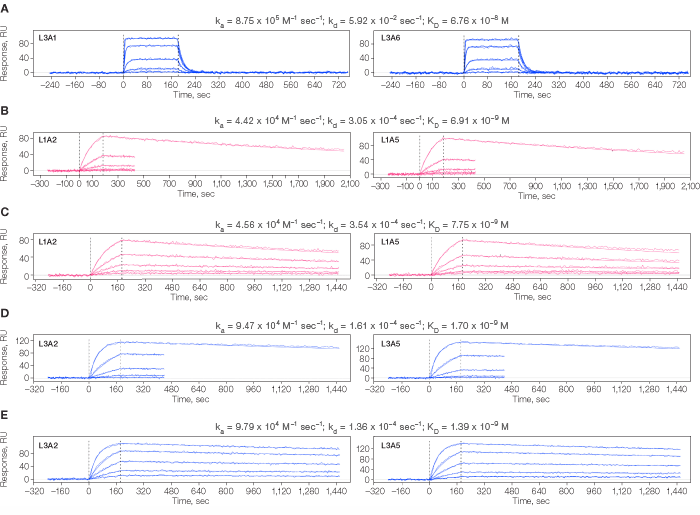
Fig 2. Kinetic analysis of analyte 1 interacting with Group I (A) and analyte 2 interacting with Group II (B–D) antibodies arrayed onto individual spots of two ProteOn chips, one per Group. Residuals not shown. A. Duplicate analyte 1 injections over the same antibody immobilized on two non-adjacent spots (L3A1 and L3A6) within the array. B. Interaction of analyte 2 binding an antibody immobilized on spots L1A2 and L1A5 using the short-and-long dissociation method. C. Interaction of analyte 2 binding an antibody immobilized on spots L1A2 and L1A5 using a long dissociation for all cycles. D. Interaction of analyte 2 binding an antibody immobilized on spots L3A2 and L3A5 using the short-and-long dissociation method. E. Interaction of analyte 2 binding an antibody immobilized on spots L3A2 and L3A5 using a long dissociation for all cycles.
Comparisons of Different Injection Methodologies
Some protein interactions in a test panel may bind stably enough to require a long dissociation time to resolve their dissociation rate constant precisely. Globally analyzing a data set that combines the use of short and long dissociation times (Katsamba et al. 2006) speeds up a kinetic analysis compared to allowing each binding cycle a long dissociation time, because both methods return similar rate constants (Figure 2B vs. 2C and 2D vs. 2E).
Diverse Kinetic Profiles
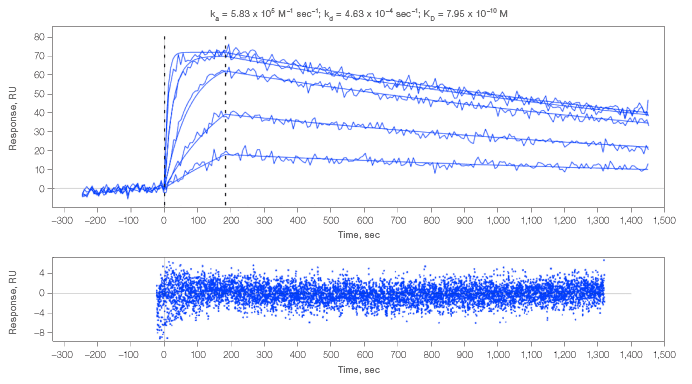
The 36-ligand array method was able to characterize kinetically diverse interactions simultaneously as shown by the data collected for the Group II panel of antibodies (Figure 2B–E and Figure 3A). Kinetic parameters were also obtained for antibodies that achieved only low levels of immobilization (Figure 3B).
A. Interaction of analyte 2 with an antibody that displayed a fast association rate
B. Interaction of analyte 2 with an antibody that coupled at a low level.
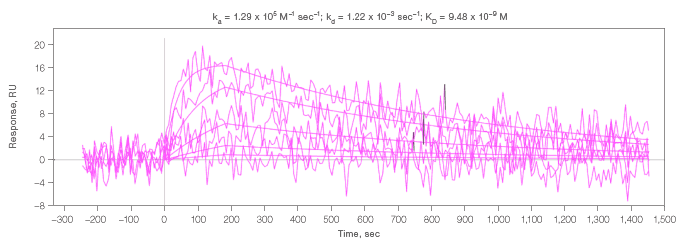
Fig. 3. Kinetic analysis of analyte 2 interacting with select spots. Residuals are close to baseline and within normal instrument noise. An example of residuals for all interactions measured in this experiment are shown in 3A.
Affinity Determination via Kinetic and Equilibrium Methods
The data for the Group I antibodies were characterized by fast kinetics and were therefore amenable to analysis via both the kinetic wizard and the equilibrium wizard. Both methods returned equivalent KD values, for example, for the interaction depicted in Figure 2A, the kinetic and equilibrium methods yielded KD values of 6.76 x 10-8 M and 6.82 x 10-8 M, respectively. An overlay of the equilibrium fits for all 36 spots is shown in Figure 4, highlighting the large amount of high-quality data that can be generated simultaneously.
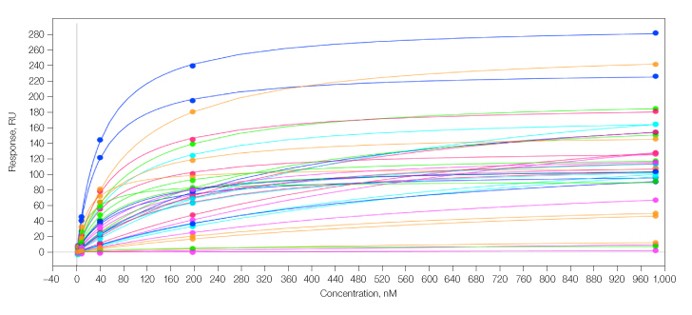
Fig. 4. Overlay of 36 equilibrium binding isotherms generated from duplicate dose-dependent analysis of analyte 1 across the Group I antibody array. In this experiment, KD values ranged from 1 x 10–6 to 1.98 x 10–8 M.
Nonspecific Binding
Nonspecific binding was evaluated on both Group I and Group II antibody arrays. Figure 5 shows that analyte 2 did not interact with an “activated and blocked” spot (without any antibody immobilized) on the Group II array.
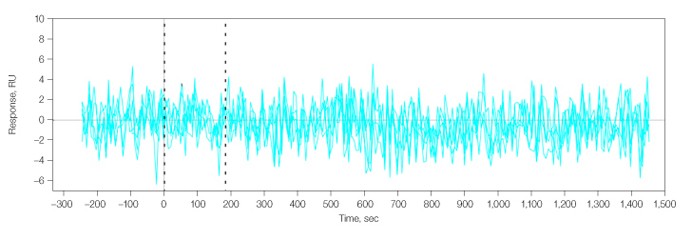
Fig. 5. No interaction was detected for analyte 2 flowed over a blank spot (L2A6), that is, without an immobilized antibody.
“Taken together, these findings gave me the confidence to apply the 36-ligand array method to expedite kinetic studies of various research projects,” says Abdiche. The impact on the team’s work in terms of increased throughput and time and cost savings has been significant. Before devising 36 array processing capabilities on the ProteOn system, Abdiche was immobilizing only six ligands per chip to perform conventional One-shot™ Kinetcs on the ProteOn system. When only six ligands are immobilized per chip, epitope binning a panel of 36 antibodies is time-consuming and necessitates the use of six chips. In contrast, the use of a 36-ligand array enables a combinatorial pairwise competition analysis of 1,296 interactions per chip by exploring a full set of 36 x 36 antibody sandwiching pairs in a single experiment. “This has meant that not only can we get our screening studies completed faster, but the ProteOn instrument has also become available sooner for other projects,” says Abdiche. “This has helped us to accelerate scientific achievement in our laboratory.”
Spreading the Word
Despite the potential for the drug discovery environment to be one that is highly competitive, Abdiche is driven to share research methods and discoveries that can help scientists elsewhere, not just in her laboratory. One of her primary goals is to affect science positively in general, not just specifically related to her projects. “Helping other researchers improve efficiency will ultimately help science move forward faster and better,” she explains. It was in this spirit of cooperation that Abdiche discussed her results with 36 x 36 array reactions using the ProteOn system with Dr John Kulman at PEPtalk in January 2010.
At the time, Kulman was heading a laboratory at the Puget Sound Blood Center in Seattle, Washington. Kulman’s lab was set up to study the biosynthesis of clotting factors and evaluate immunotherapies for thrombotic disorders. This work did not require screening hundreds of antibodies, but rather involved the detailed characterization of a few antibodies against specific clotting factors to establish mechanisms of action. The challenges in his work were therefore not related to variety of targets, but rather variety of conditions applied to the targets he studied. “I differ from many SPR researchers in that my focus has been mainly on thermodynamics,” says Kulman.
He explains that while it is common for most experiments to be conducted at a standard 25°C, his experiments were designed to test molecular interactions across a range of 10–40°C. “Evaluating interactions at each temperature change can become unruly,” he says. So the method developed by Abdiche to study 36 independent reactions in one experiment seemed the ideal solution to render these studies not only feasible, but manageable. The 6 x 6 format allowed him to do a concentration series in one, instead of multiple, injections while simultaneously evaluating several ligand variants. Kulman explains that this capability has made thermodynamic studies much more feasible than they had been in the past, reducing the time required 20-fold. “When you apply this approach to all temperature points with several ligand variants, experiments can be completed in a few days rather than a month or longer,” he explains.
Upon testing and validating Abdiche’s workflow, the limiting factor in terms of thermodynamic experimentation in Kulman’s lab was no longer time, it was scarcity of materials. “This level of throughput was not in the realm of possibility before, so we didn’t have sufficient mutants available to run the experiment at full capacity,” says Kulman. “Since we were no longer limited to four channels on the instrument, it quickly became apparent that we needed to rethink our experiments in terms of what questions we wanted to answer, not what questions the instrument was the instrument was capable of answering.”
Recently, Kulman accepted a position at Biogen Idec in Waltham, Massachusetts. Key among his responsibilities is the task of designing novel protein engineering strategies that circumvent conventional throughput limitations. “I think we will see a trend toward more high-content analysis of biomolecular interactions earlier in the development cycle, but this will depend on the integration of throughput enhancement across a temperature dimension. With the ProteOn system, this is possible right now,” he says.
Conclusions
The ProteOn protein interaction system comprises a platform that is amenable to innovation even after it has been placed in customer laboratories. Through user groups, national and international meetings, customer immersions, and collaborations, researchers have demonstrated a knack for using the system to push the boundaries of conventional protein interaction study methods. Abdiche’s development of a method for analyzing 36 simultaneous interactions is but the latest example of innovation that has already started to impact not only the efficiency and effectiveness of research in her lab, but in other SPR laboratories around the world.
References
Adibiche et al. (2011) Expanding the ProteOn XPR36 biosensor into a 36-ligand array expedites protein interaction analysis. Anal Biochem. 2011 Apr 1;411(1):139-51.
Balducci C et al. (2010). Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein. Proc Natl Acad Sci 107(5), 2295-2300.
Biasimi E et al. (2009). Immunopurification of pathological prion protein aggregates. PLoS One 4(11), e7816.
Di Fede G et al. (2009). A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 13;323(5920),1473-1477.
Katsamba et al. (2006). Kinetic analysis of a high-affinity antibody/antigen interaction performed by multiple Biacore users. Anal Biochem 352, 208-221.
Nashol O et al. (2008). Parallel kinetic analysis and affinity determination of hundreds of monoclonal antibodies using the ProteOn XPR36. Anal Biochem 1;383(1), 52-60.{jcomments on}

