Circulation of SARS-CoV-2 in the global population has led to the emergence of new variants threatening to undermine vaccine effectiveness. Understanding the immune response and its neutralizing abilities at both the patient level and globally could not only help prevent outbreaks but also mitigate the outcome for future pandemics.
The ongoing spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) over two years after the emergence of coronavirus disease 2019 (COVID-19) represents a major global health crisis for communities and hospitals while destabilizing economic growth. New variants of concern (VOCs) have emerged since the start of the pandemic due to widespread and prolonged viral circulation among the global population and ever-increasing case numbers. These variants possess evolutionary traits that may increase their transmissibility and virulence while decreasing the effectiveness of vaccines, diagnostic tests, and therapeutic tools.
To date, the World Health Organization has identified five VOCs: Alpha, Beta, Gamma, Delta, and Omicron and two additional variants of interest (VOIs): Lambda and Mu. Since the emergence of VOCs and VOIs poses a threat to both healthcare providers and policy makers managing the pandemic response, surveying the immune status of the population and monitoring evolving variants are key to ensuring a timely response to new outbreaks and evaluating the spread of COVID-19.
Monitoring Antibody Response in Asymptomatic Individuals
Since around 80% of SARS-CoV-2 infections are either asymptomatic or mild, it is not surprising that transmission by asymptomatic and presymptomatic individuals is a major contributing factor to the spread of COVID-19 (Eckerle and Meyer 2020). Therefore, monitoring only symptomatic individuals does not take into account the immune status of a large cohort of the population either currently or previously infected with SARS-CoV-2, in addition to the timing and the type of vaccines and boosters. Intelligently combining a range of surveillance tools in seroprevalence studies can help researchers understand how COVID-19 spreads in the general population while assessing the antibody response and protective immunity either from natural infection or vaccination in both symptomatic and asymptomatic individuals. This information would not only be useful for managing COVID-19 but also could help to formulate response strategies for future pandemics.
Multiplex assays, such as the Bio-Plex Pro Human SARS-CoV-2 IgA, IgM, and IgG Assays, provide data on the isotype-specific antibody response to SARS-CoV-2 nucleocapsid (N) protein, spike 1 (S1) protein and its receptor-binding domain (RBD), and spike 2 (S2) viral protein simultaneously in less than three hours. This allows researchers to measure the isotype-specific antibody response associated with different infection stages and to distinguish between natural and vaccine-induced IgA, IgM, and IgG antibody responses (Fenwick et al. 2021a).
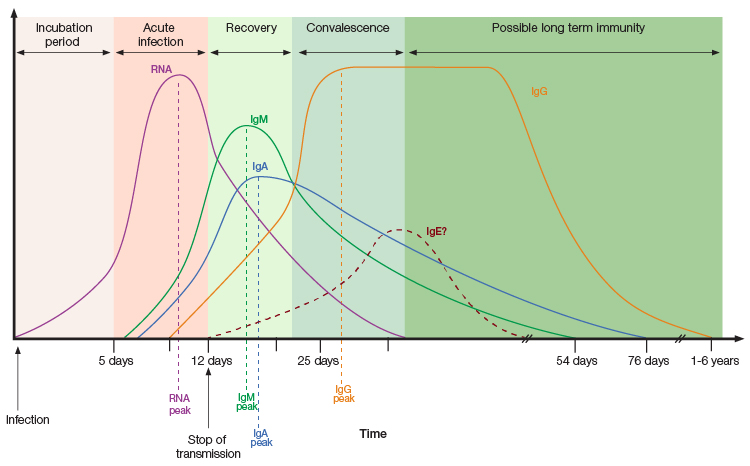
IgA is produced shortly after exposure to a pathogen, protecting epithelial barriers and modulating immune responses during inflammation. A timely IgA response to SARS-CoV-2 was found to significantly contribute towards virus neutralization, while its absence shortly after infection might be associated with severe COVID-19 disease progression, vaccine failure, and persistent viral shedding in patients with immunodeficiencies (Quinti et al. 2021). IgM levels are typically dominant in the early stages of infection, while IgG levels peak weeks after initial exposure (Liu et al. 2020). However, a delayed IgM response was observed in COVID-19 patients with severe disease progression (Shen et al. 2020). Patients with severe COVID-19 were also found to possess higher levels of IgG in later stages of the disease when compared to patients experiencing mild symptoms (Liu et al. 2020).
Therefore, serology testing can help profile the full spectrum of isotype-specific antibody responses associated with different stages of disease and symptom severity to potentially help manage symptom progression and thereby improve patient outcomes. Researchers can also use this knowledge to monitor vaccine efficacy over time and to determine the need for additional vaccine boosters in vulnerable individuals (Scurr et al. 2022).
Antibody overview and timeline in SARS-CoV-2 infection. Figure recreated from Galipeu et al. 2020 under a Creative Commons Attribution License (CC BY).
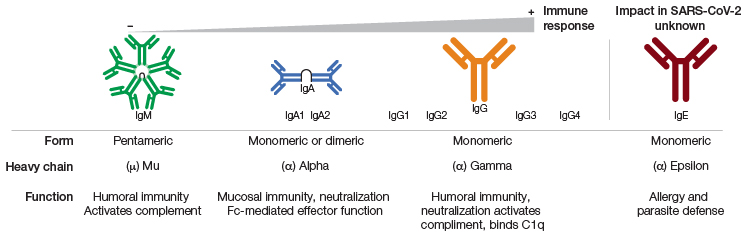
Isotype-specific antibody prevalence in patients with COVID-19. Humoral immune response generates isotype-specific antibodies at different stages of disease after exposure to SARS-CoV-2 antigens, either through vaccination or natural infection. IgA antibodies are produced shortly after SARS-CoV-2 antigen exposure generating mucosal immune response. IgM antibodies are prevalent at early stages of infection, and IgG antibodies are secreted by plasma B cells at later stages of infection. Measuring isotype-specific antibody levels can reveal the time of initial exposure, type of immunity (natural or vaccine-induced), and length of immunity. Figure recreated from Galipeu et al. 2020 under a Creative Commons Attribution License (CC BY).
Tracking Emerging Variants
Circulation of SARS-CoV-2 among the global population has facilitated the emergence of VOCs with mutations linked to significant epidemiological consequences, such as increased transmissibility, virulence, and immune response evasion. The latest VOC, Omicron, can evade immunity from both a past infection and/or two vaccine doses (Ferguson et al. 2021). This highlights the urgency for routinely monitoring variants spreading within communities. Breakthrough infections and the emergence of novel variants, capable of evading vaccine-induced immune response, could challenge and hinder current vaccination programs.
In addition to tracking variants, researchers need to understand the type and longevity of the humoral immune response. Profiling the antibody response can help differentiate between vaccinated individuals who possess only anti-spike antibodies and individuals who have been exposed to the virus naturally and may have higher antibody titers against the S1 (including RBD) and S2 subunits as well as the N protein. Neutralizing antibodies are capable of preventing viral entry into human cells by blocking the binding of SARS-CoV-2 S1 RBD to the angiotensin-converting enzyme 2 (ACE2) receptor. However, this is not the case for all antibodies, especially those that bind to variants such as Omicron with multiple mutations in the RBD subunit, enabling partial escape from antibody neutralization (Fenwick et al. 2021b; preprint: Seki et al. 2022; Cele et al. 2022).
Antibody neutralization assays, such as Bio-Plex Pro Human SARS-CoV-2 Neutralization Antibody Assays, allow researchers to monitor the efficacy of COVID-19 vaccines and compare it with the naturally occurring antibody response to emerging VOCs and VOIs. These specific assays quantitatively measure SARS-CoV-2 neutralizing antibodies against two wild-type and 11 variants of the RBD and other S1 protein subunits. In this rapidly changing SARS-CoV-2 variant landscape, the Bio-Plex Pro Human SARS-CoV-2 Neutralization Antibody Custom Assay Developer Kit can be used to develop neutralization antibody assays with any novel VOC or VOI. The results could help policy makers and public health officials make more-informed decisions on an emerging variant.
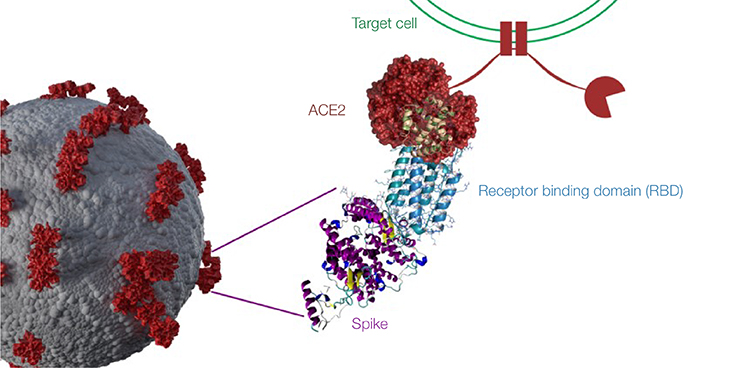
SARS-CoV-2 viral entry and neutralizing antibody mechanism. Neutralizing antibodies to SARS-CoV-2 block virus entry into host cells by binding to part of the virus’s spike protein. When the virus spike entry is blocked, infection is prevented. The spike protein subunit S1 contains the receptor binding domain (RBD) — a prime target for neutralizing antibodies because of its role in viral entry into host cells by binding to the host ACE2 receptor. Mutations in the RBD can potentially result in SARS-CoV-2 variants that are more infectious with increased transmissibility or reduce the effectiveness of current vaccines. ACE2, angiotensin-converting enzyme 2.
Keeping Up with Emerging Variants
At this stage of the postvaccine pandemic, the scientific response has moved to monitoring novel SARS-CoV-2 variants and identifying risk factors associated with increased transmissibility, virulence, and immune evasion. To stay ahead of new variants, policy makers and public health officials must focus on community-level testing to ensure vaccines remain effective and to detect outbreaks in a timely manner. A comprehensive surveillance platform will help evaluate the spread of SARS-CoV-2 within a community, allowing governments to stay on top of the pandemic and respond more efficiently to any future pandemics that may arise.
References
Cele S et al. (2022). Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602, 654–656.
Eckerle I and Meyer B (2020). SARS-CoV-2 seroprevalence in COVID-19 hotspots. The Lancet 396, 514–515.
Fenwick C et al. (2021a). Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J Virol 75, e01828–20.
Fenwick C et al. (2021b). A multiplexed high-throughput neutralization assay reveals a lack of activity against multiple variants after SARS-CoV-2 infection. medRxiv. Preprint. https://doi.org/10.1101/2021.04.08.21255150, accessed March 24, 2022.
Ferguson N et al. (2021). Growth, population distribution and immune escape of Omicron in England. Imperial College London. https://doi.org/10.25561/93038, accessed March 24, 2022.
Galipeau Y et al. (2020). Humoral responses and serological assays in SARS-CoV-2 infections. Front Immunol 11, 610,688.
Quinti I et al. (2021). IgA antibodies and IgA deficiency in SARS-CoV-2 infection. Front Cell Infect Microbiol 1, 655,896.
Liu X et al. (2020). Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect 9, 1,269–1,274.
Scurr MJ et al. (2022). Whole blood-based measurement of SARS-CoV-2-specific T cells reveals asymptomatic infection and vaccine immunogenicity in healthy subjects and patients with solid-organ cancers. Immunology 165, 250–259.
Seki Y et al. (2022). Safety and immunogenicity of Pfizer/BioNTech SARS-CoV-2 mRNA third booster vaccine against SARS-CoV-2 Omicron variant in Japanese healthcare workers. medRxiv. Preprint. https://doi.org/10.1101/2022.01.20.22269587, accessed March 24, 2021.
Shen L et al. (2020). Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg Microbes Infect 9, 1,096–1,101.
Variants of Concern, World Health Organization. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/, accessed March 24, 2022.

