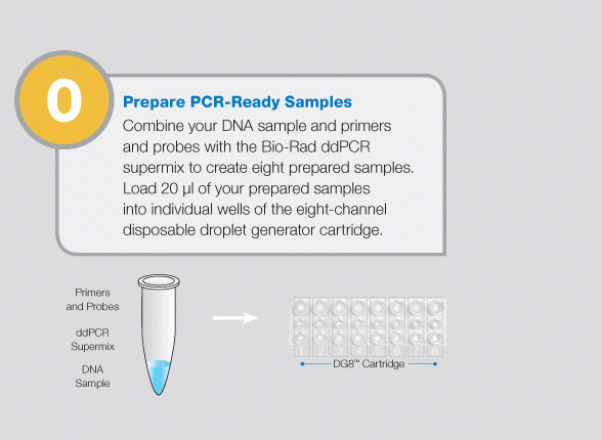
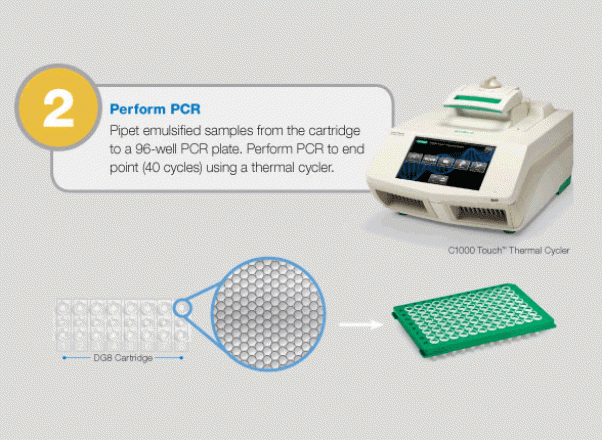
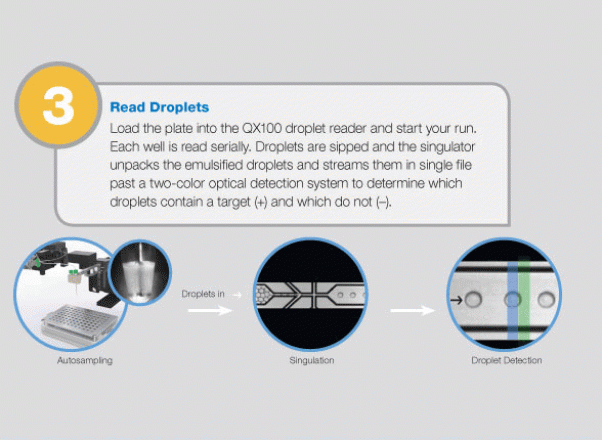
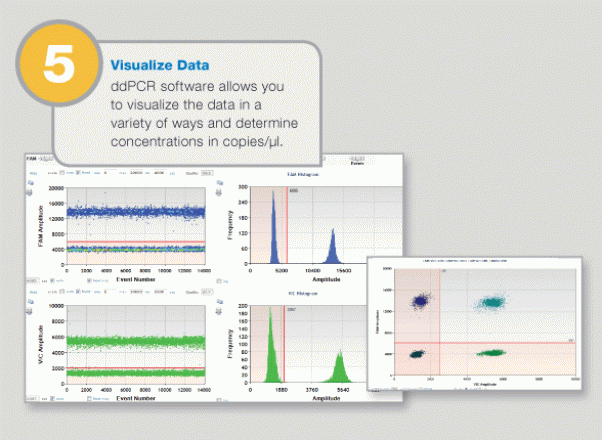
Large-Scale Genomic Variation Research
“People are different from each other in ways that are fascinating and medically important, and to understand the ways in which variations in our genome give rise to those differences presents an important and interesting set of questions,” says Dr Steve McCarroll, faculty member and principal investigator with the Department of Genetics at Harvard Medical School and the Broad Institute of Harvard and MIT. McCarroll, who came to human genetics by way of C. elegans genetics (he did his PhD in Dr Cori Bargmann’s laboratory, then his postdoctoral work with Dr David Altshuler), says he was drawn to the challenge of using human genome variation to identify the genes underlying biological processes and human disease.

Faculty member and principal investigator,
Department of Genetics
Harvard Medical School and the Broad Institute of Harvard and MIT.
The McCarroll laboratory studies the biological effects of human genome polymorphism, seeking to define how genome variation influences gene expression and risk of disease. About half of his lab works to understand genome variation in general — its distribution, molecular properties, and influence on gene expression — the other half works to use genome variation to reveal the genetics underlying schizophrenia and bipolar disorder.
McCarroll explains that genomes vary in different ways and at different scales. Single nucleotide polymorphisms (SNPs) are at the small end of the scale with one-letter (single nucleotide) differences in genetic codes. Since the late 1990s there has been a significant effort to understand how these single nucleotide variations might lead to disease. But it wasn’t until the mid-2000s that the technology to study the other end of the spectrum — genome segments tens to hundreds of thousands of base pairs long that are present in different numbers of copies in different people — began to emerge. “We work to understand the extent to which that large-scale variation exists, how it relates to human population genetics and population history, and how it influences human phenotypes,” says McCarroll.
Studying these regions requires being able to quantify how many copies of a particular genomic segment are present in each person’s genome. This requires the ability to measure high copy segments in large cohorts accurately, specifically, to be able to measure the copy number of a genomic sequence with integer precision in hundreds of people. That’s one of the core experimental efforts conducted in the McCarroll laboratory, an effort that has proved one of the greatest challenges to furthering their research. “We tried various things over the years,” says McCarroll. “Using technology such as real-time PCR and CGH (comparative genomic hybridization) arrays, we could measure simple deletions and duplications, such as those that change the copy number of a segment in a person’s genome from two to one or zero, or from two to three or four. But what we lacked was the ability to measure copy numbers greater than four in a way that was highly precise and reproducible.”
This measurement is precisely what Droplet Digital™ PCR (ddPCR™) technology was developed to enable researchers to do.
ddPCR Technology Defined
Representing third-generation PCR technology, ddPCR is an absolute measure that enables single-molecule resolution of target sequences with extreme precision and accuracy. This system uses microfluidics to partition samples into 20,000 individual nanoliter droplets, each a separate PCR reaction. The QX100™ Droplet Digital™ PCR system provides a revolutionary approach to target DNA quantification.
The QX100 system consists of two instruments: the droplet generator and the droplet reader. The droplet generator partitions each sample to 20,000 individual nanoliter droplets. The droplets are transferred into PCR plates and moved to a standard thermal cycler, where the targeted DNA/RNA molecules are amplified. The PCR plate is then placed in the droplet reader, where the droplets are streamed, single-file, past a two-color LED detector that reads each droplet as either positive or negative for the target DNA/RNA molecules. The system software then determines the concentration of the selected target in the original sample and provides absolute quantification in digital form. The system allows detection of slight differences in gene copy number (e.g., six copies from five) or a 10% difference in mRNA expression with 95% confidence. Since actual molecules are being measured, there is no need for a standard curve or reference gene.
The precision of the QX100 ddPCR system enables small-fold (1.2x) differences, making it ideal for researchers seeking to perform high-resolution copy number variation (CNV) (as is the case the McCarroll lab). The QX100 can also be used for rare event detection (up to one mutant copy of an SNP (BRAF V600E) in 100:000 wild-type copies), and absolute quantification of nucleic acids without the need for a standard curve. The system is also ideal for gene expression studies, including miRNA amplification and mRNA expression analysis for a number of genes.
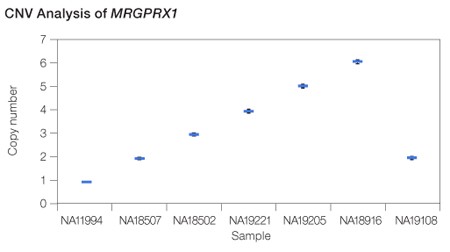
Calculation of copy number variation. For MRGPRX1, the copy number states from 1 to 6 were completely resolved.
Impact of ddPCR in the McCarroll Lab
McCarroll first heard about ddPCR as a post doctoral scientist in 2007, when an early version of the technology was described by researchers in the journal Analytical Chemistry. So when the first products were introduced to the market in mid-2011, McCarroll’s lab was one of the first to acquire the QX100 system. Immediately, researchers in his laboratory began incorporating the instrument in ongoing projects to study regions of the genome that are structurally complicated (regions that have been influenced by many different structural mutations in their history). In such regions, this technology has allowed them to quantitate copy numbers in large populations, which is important in both population genetic analysis and disease analysis. It has also allowed them to begin to gain insight into how these structurally complicated regions relate to human phenotypes. “It lets us see how the copy number of a genomic sequence varies within a population, what parts of the world it varies in, and how variation in it relates to other genetic variations in that region of the genome,” says McCarroll. “Then we can also, most importantly, measure the copy number of a genomic sequence in case-control cohorts and evaluate how the copy number in that region relates to risk of disease.”
Potential uses for ddPCR
Studies incorporating ddPCR technology in laboratories across the globe include:
- Absolute quantification of HIV proviral DNA and human genomic DNA in the same reaction
- Absolute quantification of HIV viral RNA using two-step reverse transcription-ddPCR
- Rare event detection of BRAF V600E, EGFR L858R and T790M, JAK-2, and cKIT (cancer mutation analysis and drug resistance mutation)
- Absolute quantification of miRNA from circulating nucleic acids for biomarker identification
- High-resolution copy number variation (CNV) analysis of human and mouse DNA’s
- High-resolution CNV analysis of the MET gene, association study for lung cancer
- Absolute quantification of next generation sequencing library
- Rare event detection, mitochondrial mutagenesis
- Absolute quantification of the BCR-ABL (Philadelphia chromosome) fusion gene and transcript, association with leukemia
- Absolute quantification and sex determination of fetal DNA in circulating nucleic acid purified from maternal blood
McCarroll explains that until ddPCR technology was developed, these regions in the genome could not be analyzed in the disciplined, high-quality, high-precision way that is the standard for genetic analysis of simpler kinds of variation, such as SNPs. Because of this challenge, his lab avoided studying these regions since the quality of data that could be generated with more conventional technologies didn’t meet the standards on which his lab wanted to base scientific inference and analysis. “The ddPCR technology now allows us to go straight into these regions and start figuring them out,” says McCarroll.
Since the lab acquired it, ddPCR technology has begun to play an important role in three research projects there. In addition to the studies of structurally complex regions, the McCarroll lab has been using ddPCR to understand genetic influences on gene expression that affect risk of schizophrenia and bipolar illness. The QX100 platform is being used to obtain crisp, precise measurements of gene expression, giving researchers the power to analyze how expression of each gene correlates with genome variation in regulatory regions of the gene across large sets of tissues from different individuals.
“These higher, multi-allelic copy number variations that we’re now able to study with this technology contribute substantially to human genome variation,” says McCarroll. “They influence the copy number of hundreds of human genes, but we’ve never really had the tools to study them accurately in human genetics. Now that we can study them in precise, disciplined ways, it’s opening a lot of scientific doors.”
Droplet Digital PCR Workflow Overview
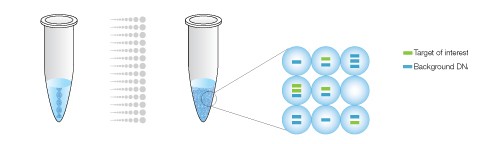
The sample is partitioned into 20,000 droplets, with target and background DNA randomly distributed smong the droplets.
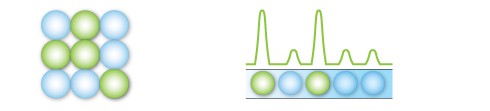
After PCR amplification, each droplet provides a fluorescent positive or negative signal indicating the target DNA was present or not present after partitioning. Each droplet provides an independent digital measurement.
![]()
Positive and negative droplets are counted for each samples, and the software calculates the concentration of target DNA as copies per microliter.