Clinical researchers in Sweden use sensitive Droplet Digital PCR Assays to detect blood cancer relapse early, enabling early treatment intervention for better patient outcomes.
No patient who has survived cancer wants to hear that they’ve relapsed. But hearing it sooner rather than later can dramatically increase the chances of finding a successful, potentially life-saving treatment.
Myelodysplastic syndrome (MDS) is a type of cancer in which bone marrow stem cells mature improperly. The disease can develop into acute myeloid leukemia (AML), a more serious type of blood cancer. For severe cases of MDS, the recommended treatment is chemotherapy followed by allogeneic stem cell transplantation. Unfortunately, even with this advanced treatment, which is difficult and can be painful, more than 30% of patients relapse, and the cancer has often progressed too far to halt by the time relapse is detected clinically.
One group of scientists and clinicians has set out to change that. The Nordic MDS Study Group is currently in the first phase of a study enrolling patients with MDS undergoing allogenic stem cell transplantation, where they are applying Droplet Digital PCR (ddPCR) technology as a sensitive, efficient tool for monitoring patient-specific molecular biomarkers that signal the presence of minimal residual disease (MRD) that could cause relapse. By carrying out regular monitoring to detect even extremely low levels of resurgent cancer, this strategy gives doctors the chance to intervene at a stage when treatment could still make a difference for patients. During the second phase, MRD will be delivered in real-time and be used to initiate clinical intervention.
Validating a New Diagnostic Workflow
Current methods for detecting relapse in MDS and related diseases rely primarily on cell morphology using methods like flow cytometry. It can be challenging to detect cancer cells with these methods when there are only a few lingering in the body — even though that is the optimal time to eradicate them. Next-generation sequencing (NGS) is a comprehensive way to find mutations. But it is not an efficient tool for regular, ongoing disease monitoring due to its expense and the volume of data it generates.
The Nordic MDS researchers came up with a multistep workflow that leverages all of the most sensitive technologies efficiently. Following diagnosis, the researchers sequence samples from patients with MDS using a myeloid gene panel for next-generation sequencing (NGS) to identify their unique disease mutations. Then, after stem cell transplantation treatment, the researchers take blood and bone marrow samples every three months and run detection assays for the relevant mutations using the Bio-Rad QX200 Droplet Digital PCR System. The study group has also collected those samples into a biobank that they hope will eventually yield additional insights into MDS as a disease — its progression, its genetic contributions, and factors that influence treatment outcomes.
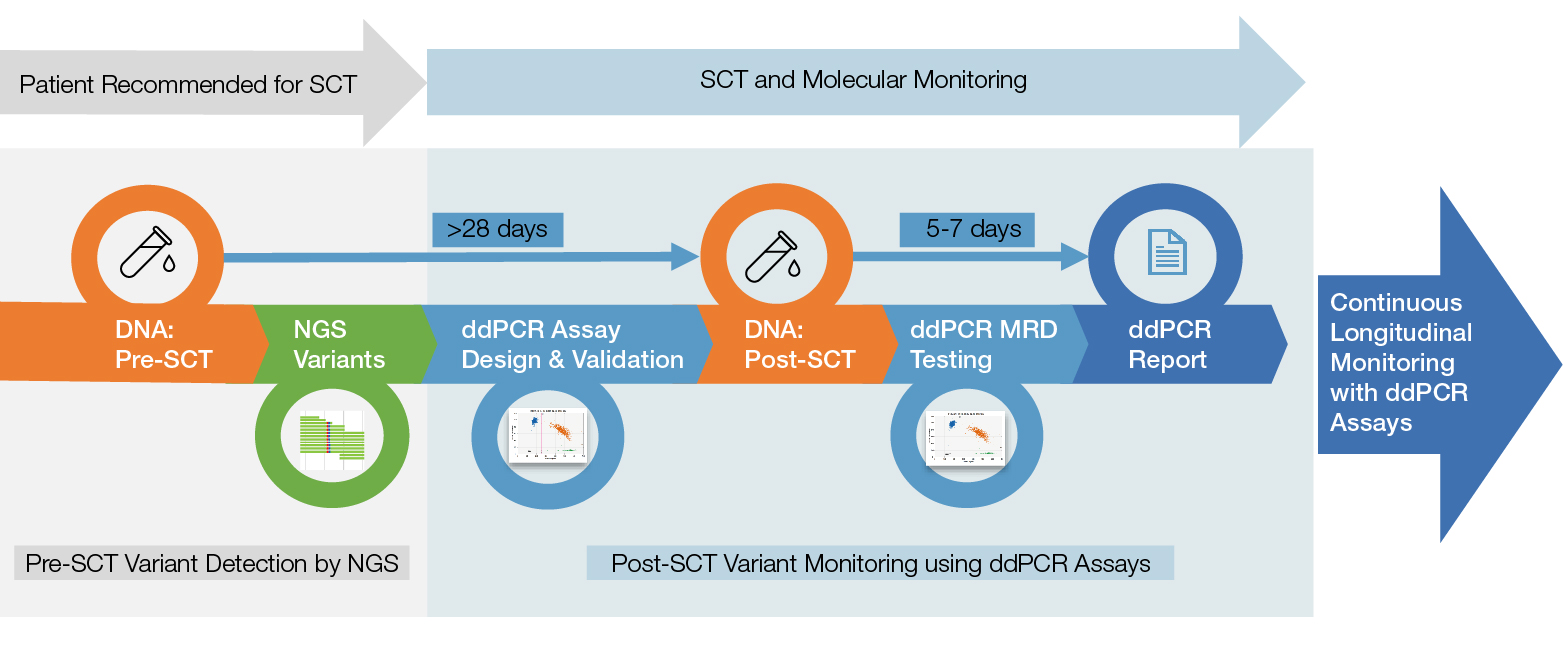
From stem cell transplant to molecular monitoring with Droplet Digital PCR (ddPCR). Patients are screened for relevant mutations using next-generation sequencing (NGS) prior to stem cell transplant (SCT). Multiple individual ddPCR Assays are designed for each patient and are used to monitor for post-transplant relapse every three months for up to 24 months. MRD, minimum residual disease.
“The low cost of ddPCR Assays allows us to monitor each patient frequently, which is very important,” says Lucia Cavelier, PhD, an Associate Professor in Medical Genetics at the University of Uppsala and one of the study’s lead designers and genetic analysts.
Thus far, the study has sequenced samples from over 300 patients, identifying 690 significant genetic variants and designing over 350 unique ddPCR Assays to monitor those variants following stem cell transplantation. The researchers defined a sensitivity threshold of 0.1% across all assays to facilitate validation. However, they noted that many of the ddPCR Assays demonstrated much better sensitivity, with an apparent false positive rate of zero.
“We’ve come to trust the ddPCR data very highly,” says Simone Weström, PhD, a senior research engineer at the University of Uppsala who has worked on the project. “In cases where there is a disagreement between ddPCR results and confirmatory NGS data, it has usually been explained by some sort of allelic dropout or a problem with the sequencing.”
While the current project involves collaboration between clinical institutions and academic laboratories, the researchers say the ddPCR Assays are easy to use and could be applied by clinicians without academic intermediaries. The protocols are relatively simple, and data analysis doesn’t require specialized software.
“We chose to use ddPCR technology because of its reliability and precision,” says Cavelier. “This program has allowed us to validate the method, and I think it will be used in many labs and clinics across Sweden and beyond. It’s relatively cheap, fast, flexible, and highly reproducible. It’s also simple — it just gives you a positive or negative result about the presence of the mutation without adding unnecessary information. That’s what makes it a good clinical assay.”
Making a Difference for Clinicians and Patients
The ultimate goal of this study is to lower rates of full-blown MDS relapse by allowing clinicians to intervene with chemotherapy and other treatments while there are relatively few cancer cells in a patient’s body. Treatments at this stage are much more likely to be successful, giving transplanted stem cells time to grow and replace cancerous cells. “The survival rate for these diseases is very low. So if we can avert relapses early, we can give patients a lot of years they otherwise wouldn’t have,” says Cavelier.
Even for patients who don’t relapse, the sensitivity of ddPCR Assays can give added peace of mind. “Imagine if you have a child with leukemia — if you can know with high certainty that [their] sample is negative, that’s amazing for parents,” says Cavelier. “For doctors, it’s great to be able to say, ‘We don’t see it, it’s not there; and if we do start seeing it, we’ll be able to change treatment right away.’”
The Nordic MDS Study Group researchers foresee ddPCR Assays becoming a more regular part of clinical practice, offering patients and doctors information on a molecular level that can deepen understanding and inform treatment decisions.
“We were pleasantly surprised by just how enthusiastic clinicians have been about the Droplet Digital PCR Assays,” says Cavelier. “Doctors here are very cost sensitive, and these assays are relatively cheap compared to many other methods while still giving excellent information. People also like that the results are fast and simple. No one wants to wait around for a large dataset full of new mutations — they just want to know whether the mutations of interest are there or not. So I think interest in ordering these tests is quite high in the medical community.”
Learn more about the benefits and applications of Droplet Digital PCR.
References
Nordic MDS Group (n.d.). NMDSG14B: Individual molecular minimal residual disease monitoring for patients with MDS and mixed MDS/MPN treated with allogeneic stem cell transplantation. https://www.nmds.org/index.php/clinical-trials/73-nmdsg14b. Accessed May 21, 2021.

