Be it for producing vaccines or isolating biomarkers, protein purification is an important staple of the pharmaceutical industry. Proteins are purified to characterize and isolate desired functions, structures, or interactions. They can be purified using a range of techniques, but the most common is chromatography. Chromatography relies on a stationary phase (typically resin) to separate desired proteins based on characteristics such as hydrophobicity, size, chemical affinity, and even charge. Often, more than one parameter is needed to isolate a target protein to desired purity, resulting in multistep processes. This means that, whether in a bench setting or on an industrial scale, isolating a target protein can be laborious and time consuming. One way to minimize this is to use mixed-mode resins — resins that separate based on more than one parameter at once, thus allowing for differential purification of proteins with otherwise similar characteristics.
For the past few decades, mixed-mode resins have been used to improve protein purification techniques since they offer higher selectivity and efficiency than their single-interaction counterparts. One example of a mixed-mode resin is Bio-Rad’s Nuvia™ cPrime™ Resin. Nuvia cPrime, designed for a wide variety of proteins, has a ligand with three functional groups: a weak carboxylic acid, an aromatic hydrophobic ring, and an amide bond (Figure 1). Nuvia cPrime Resin was also designed for superior flow properties, mass transfer, and stability, making it ideal for process-scale purification. However, due to the covariate nature of mixed-mode purification, designing process optimization experiments regularly adds an extra level of complication.
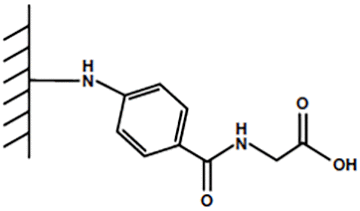
Fig. 1. Mixed-mode ligand for Nuvia cPrime Resin.
To mitigate the inherent complexity in using mixed-mode resins, design of experiments (DoE) is employed. DoE, often carried out using dedicated software, is a helpful method for determining connections between process parameters (like pH or salt concentration) and purification outcomes. The major advantage of using DoE is that critical process relationships do not have to be determined empirically. By evaluating multiple parameters at once, DoE can also reveal important process relationships that may not be understood when considering single input-outcome association alone. This means DoE can take much of the stress out of optimizing purification processes.
To investigate how practical DoE is with Nuvia cPrime Resin, parameters such as salt concentration and pH were chosen to study effects on binding and elution for the target protein (pI = 5.5; MW = 51 kD; GRAVY score = -0.275). Parameter effects were evaluated via a custom matrix design with JMP Software including three center points and a total of 24 experiments. With a minimal amount of protein and resin, a simple DoE setup was used to study the selectivity, recovery, and robustness of Nuvia cPrime Resin. Conditions established in scaled-down studies like this one can also be used for process-scale purification. Watch our on-demand webinar to view a demonstration of our DoE approach using Nuvia cPrime Resin.
Learn more about Nuvia cPrime Resin and how it can help improve your protein purification process.
JMP is a trademark of SAS Institute, Inc.

