The EpiQ chromatin analysis kit is a novel epigenetics research tool for the quantitative assessment of chromatin structure in cultured cells. By combining in situ chromatin digestion, genomic DNA purification, and real-time PCR, the chromatin state for several gene promoters can be studied simultaneously. The EpiQ kit helps quantify the impact of epigenetic events such as DNA methylation and histone modification on gene expression regulation through chromatin state changes. Genes that are actively transcribed are associated with “open” or “accessible” chromatin regions, while genes that are transcriptionally silent are often in “closed” or “inaccessible” chromatin regions (Figure 1).
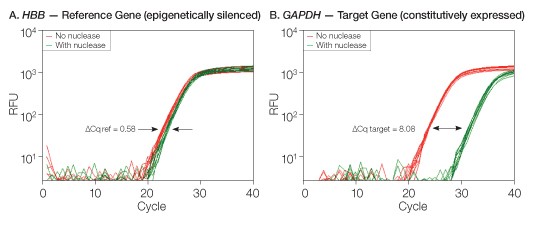
Fig. 1. The EpiQ chromatin analysis kit utilizes nuclease accessibility to discriminate open vs. closed chromatin regions. Amplification of proximal promoter regions for the epigenetically silenced HBB (reference) gene or the constitutively expressed GAPDH (target) gene was carried out in HeLa cells using the EpiQ kit and EpiQ™ chromatin SYBR® Green supermix on the CFX96™ real-time PCR detection system. A, closed chromatin regions resulted in minimal quantification cycle (Cq) delays (∆Cq = 0.58) following nuclease treatment; B, open chromatin regions led to significant Cq delays (∆Cq = 8.08) after nuclease treatment. RFU, relative fluorescent units.
The EpiQ chromatin analysis kit discriminates between open and closed regions by employing a nuclease accessibility assay and real-time PCR to quantify the level of accessibility. Rapid results can be obtained in approximately six hours and strongly correlate with gene expression levels (Figure 2). The EpiQ kit complements existing epigenetic assays (DNA methylation and histone analysis using chromatin immunoprecipitation), and researchers worldwide are using the kit to obtain novel insights into mechanisms of gene regulation.
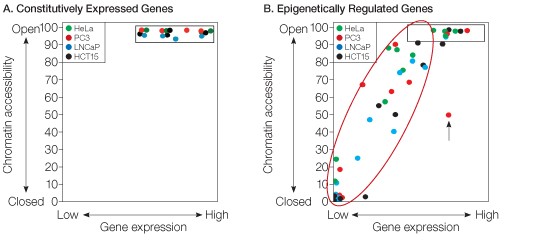
Fig. 2. Chromatin structure data from the EpiQ chromatin analysis kit strongly correlate with gene expression levels for several genes in various cell lines. A, the chromatin state of the proximal promoter regions for four constitutively expressed genes (GAPDH, ACTB, TBP, and B2M) was assessed in four different cell lines (HeLa, PC3, LNCaP, and HCT15). Each gene promoter was determined to be in an open chromatin structure. B, 11 epigenetically regulated gene promoters (ABCB1, CDH1, CDH13, DAPK1, DKK1, GSTP1, PTGS2, TP73, WT1, p14ARF, and p16INK4A) were evaluated for chromatin structure in the same four cell lines. Chromatin state varied across the panel of genes and strongly correlated with the gene expression levels. Promoter regions in an open state were associated with medium to high levels of gene expression (shown in the black box, upper right), whereas those in a closed state were linked to low expression (lower left).
Making Connections in Cancer Cell Proliferation

Department of Structural and Functional Biology
University of Insubria, Varese, Italy
Together with a group of Japanese researchers, Dr Ian Marc Bonapace discovered the UHRF1 protein in the late 1990s. At that time, Bonapace was searching for proteins involved in the control of cellular differentiation and proliferation. Since its discovery, UHRF1 has been shown to play an important role in gene regulation; specifically, in controlling the epigenetic machinery that reinstalls correct modifications on DNA and chromatin during cell proliferation.
The main objective of Bonapace’s studies is determining ways UHRF1 can ultimately affect cancer treatment. “We believe at minimum, that this protein can be very useful as a marker for cancer aggressiveness,” explains Bonapace. “But what we hope to discover in the future, is that hampering the activity of UHRF1 could be a way to stop tumor growth and metastasis in the body.”
Some of the primary techniques used in this research include chromatin immunoprecipitation, protein extraction and purification, and coimmunoprecipitation and gel filtration to identify the complexes in which this protein is involved. Early in 2010, Bonapace obtained the EpiQ kit. To test its efficacy and applicability to his overall research efforts, he conducted an experiment to compare the expression of three different oncosupressors in two different lines of prostate cancer (LNCAP, a line that is not aggressive, and PC3, a line that comes from a very aggressive tumor). “The results of these tests showed what we expected to see based on previous research,” says Bonapace. “The oncosuppressors were not downregulated in LNCAP cells, but completely downregulated (so chromatin was in a closed state) in PC3 cells.”
Bonapace explains that prior to obtaining the EpiQ kit, determining whether genes are being expressed or not required days of work using the more resource-intensive chromatin immunoprecipitation method. “With this kit,” explains Bonapace, “you can determine immediately if genes are repressed. Then you know whether or not to go through the more time-consuming and money-consuming experiments to study why.” Bonapace describes the potential impact of EpiQ kit technology on his research as providing significant time savings and efficient use of resources as he continues to test different kinds of cells and tumors to see if the same genes of interest are or are not controlled by the presence or absence of the UHRF1 protein. “The main application of this kit is not so much to aid in the discovery of something,” says Bonapace, “but more to provide a shortcut to understanding if a gene is expressed or not. Once you’ve understood that, you can go on to design studies to define better and in more detail why.”
Determining the Mechanisms of HIV Latency

Graduate Student
University of California, Berkeley
When Siddharth Dey first came to the University of California, Berkeley for graduate school, he came across a paper published in Cell by researchers in the Schaffer lab that proposed a novel model of HIV latency. The paper demonstrated that the stochastic fluctuations in a virally encoded protein that binds to a viral RNA hairpin to activate transcription, thus forming a positive feedback loop of transcription, could influence the viral latency decision. Initially, the number of these viral molecules is very low within the cell. The new model proposed that there are two options for whether a viral protein can bind to the RNA hairpin and drive transcription, thus giving rise to proliferation of the viral protein. It either starts a positive feedback loop of transcription and activates the virus, or the initial low numbers of the viral protein fail to activate the positive feedback loop, rendering the virus latent. The latent viruses are not susceptible to antiretroviral drugs, and the positive feedback loop can activate at a later time, making it impossible to completely eradicate the virus from infected patients. It was this new model of HIV latency that piqued Dey’s interest in studying the evolution of these viral proteins and made him decide to devote his graduate studies to work in the Schaffer lab.
Since joining the lab, Dey has been working on the evolution of HIV. “HIV is divided into various subtypes,” says Dey, “and it has a very high error rate, so basically you have this huge diversity. One of my goals is to understand how this diversity affects function, and how the virus still manages to operate so efficiently in spite of the variability: are there some underlying structural factors that are important for protein function?”
Dey is also interested in applying research that has helped characterize strains of HIV that infect people in the U.S. or Western Europe to strains that infect other world populations that are far less studied. “I started looking into how these different processes that got me first attracted to the lab can vary when you have different sequences,” explains Dey. “Essentially, how does sequence diversity within this protein affect various pathways or gene expression of the virus?”
In recent experiments, Dey studied a viral protein that has been shown to exhibit significant posttranslational modification and interact with a variety of cellular proteins. Several conserved sites within the protein have been identified as critical for the protein to function and if these are mutated, the virus becomes nonfunctional. “I was interested in looking one step ahead and identifying potentially nonconserved sites that may be functionally important,” explains Dey. “You can have situations where not just one site is conserved, but two sites simultaneously are conserved, so sites can co-evolve. In other words, amino acids at certain sites are correlated with amino acids at other sites.”
Part of this work required applying statistical methods on sequence alignments of the protein to identify sites that are co-evolving. Once potential sites were identified computationally, Dey was able to test these sites experimentally. “These sites definitely looked like they were co-evolving, which told us that if you just look at a sequence alignment, they may not look functionally important on a site-to-site basis, but there’s definitely correlation between the sites,” explains Dey. “Once we identified these co-evolving sites and verified them experimentally, the next step forward was to ask: how are they important either structurally or functionally?” Further experimentation demonstrated functional attributes that these co-evolving sites satisfy. If the co-evolving pairs are disrupted, the viral protein loses its function.
The EpiQ kit played a central role in these functional experiments. The viral protein has been shown to interact with many cellular proteins, including SWI/SNF, a chromatin remodeling complex. “So I attempted to determine how mutations within the viral protein affect the way in which it interacts with the chromatin remodeling factor,” explains Dey. “The EpiQ kit made it possible for me to evaluate the chromatin structure around the integrated virus, and whether the viral protein affects it.” In the past, Dey used chromatin immunoprecipitation to address similar questions. For the purposes of this and his ongoing research, the EpiQ kit does not replace this technology, rather it complements it, giving insight into the circumstances under which further analysis using chromatin immunoprecipitation is warranted.
Dey is hopeful that the questions being asked in the Schaffer lab related to HIV will eventually lead to insights into why HIV might be present in a person who does not exhibit active signs of disease. “Ultimately,” explains Dey, “we’d like to be able to completely understand what is happening at the molecular level in terms of HIV latency, so mechanisms can be developed to activate the virus completely, then apply an antiretroviral drug to kill it off.”
“The EpiQ kit made it possible for me to evaluate the chromatin structure around the integrated virus, and whether the viral protein affects it.”

