A recent publication from researchers at Columbia University Irving Medical Center (New York, NY) shows that expression of a protein involved in homology-directed repair (HDR) can improve CRISPR-Cas9 gene editing efficiency (Nambiar et al. 2019). Manipulation of DNA repair pathways and improvements to gene editing workflows, such as the addition of cell sorting, can facilitate new applications of CRISPR-Cas9 technology.
Inefficiency Hinders CRISPR-Cas9–Assisted Gene Editing
The CRISPR-Cas9 system uses a programmable guide RNA and associated Cas9 nuclease to create double-stranded breaks at targeted regions of the genome. These double-stranded breaks are typically repaired by one of two innate DNA repair pathways: non-homologous end-joining (NHEJ) or homology-directed repair (HDR). NHEJ, the dominant pathway in normal cells, is error-prone, often resulting in short insertions or deletions, which may produce a frameshift. Although repair via NHEJ can be useful for creating gene knockouts, its unpredictable nature limits its practical applications. In contrast, HDR can be used for precise deletions, mutations, and insertions, including insertions of entire genes. This is possible because HDR utilizes a template for DNA repair, which can be exogenously supplied by the researcher. However, the rarity of HDR repairs makes typical gene editing experiments highly inefficient.
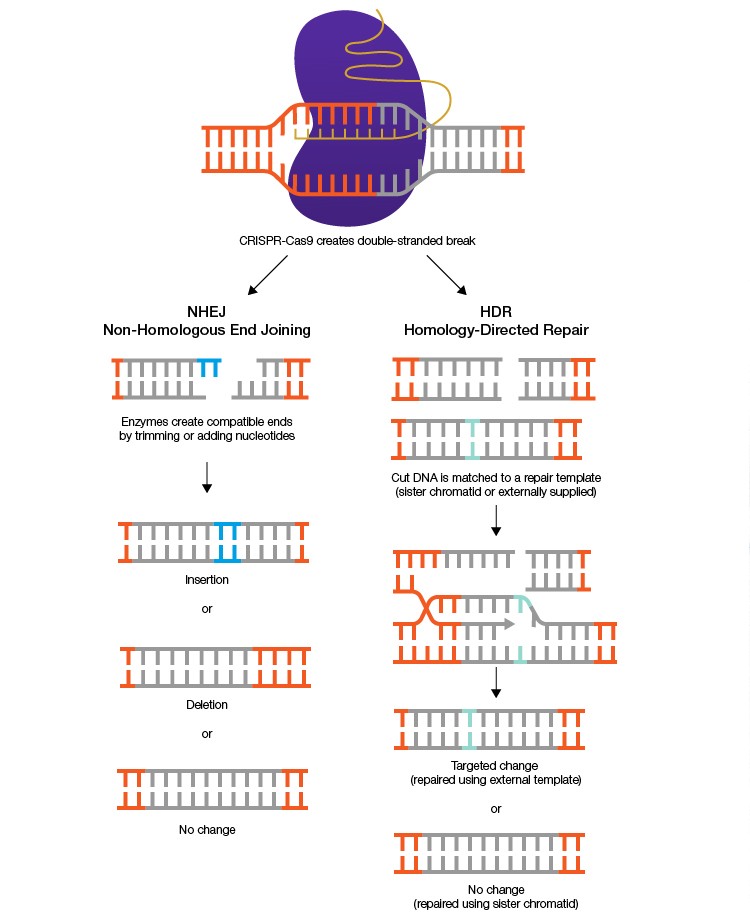
Expression of RAD18 Boosts Gene Editing Efficiency
In a 2019 publication in Nature Communications, authors Tarun Nambiar and colleagues detail a method for improving gene editing efficiency via HDR. Using fluorescent protein expression as a marker for successful HDR-dependent gene editing, researchers screened 204 ORFs encoding proteins related to DNA repair or replication. Expression of RAD18, a ubiquitin ligase involved in HDR and post-DNA replication repair, was found to have the greatest positive effect on HDR efficiency, regardless of donor template type. This effect was consistent across all three tested human cell lines. Using structural and domain-swap analyses, researchers were able to modify the expressed RAD18 sequence to further improve its effect.
The inherent inefficiency of HDR-dependent CRISPR-Cas9 gene editing leads to a waste of resources, as researchers must culture, transfect, and screen large numbers of cells to find the small proportion containing the desired edit. Methods that improve efficiency, such as RAD18 expression, are needed to help fully realize the potential of CRISPR-Cas9 technology.
The S3e Cell Sorter Simplifies CRISPR-Cas9 Gene Editing Workflows
The low efficiency of traditional HDR-dependent gene editing protocols means that the majority of the cells recovered post-transfection do not contain the desired gene edits; therefore, an enrichment step for a selectable marker is often included. Researchers in this study used a Bio-Rad S3e Cell Sorter to isolate edited cells expressing fluorescent marker proteins. Using an S3e Cell Sorter for enrichment helps preserve cell health by reducing passaging and can speed up gene editing workflows by ~30 days. Cell sorting also improves capture of rare edits and can be used for some cell types which are not compatible with traditional antibiotic resistance-based selection.
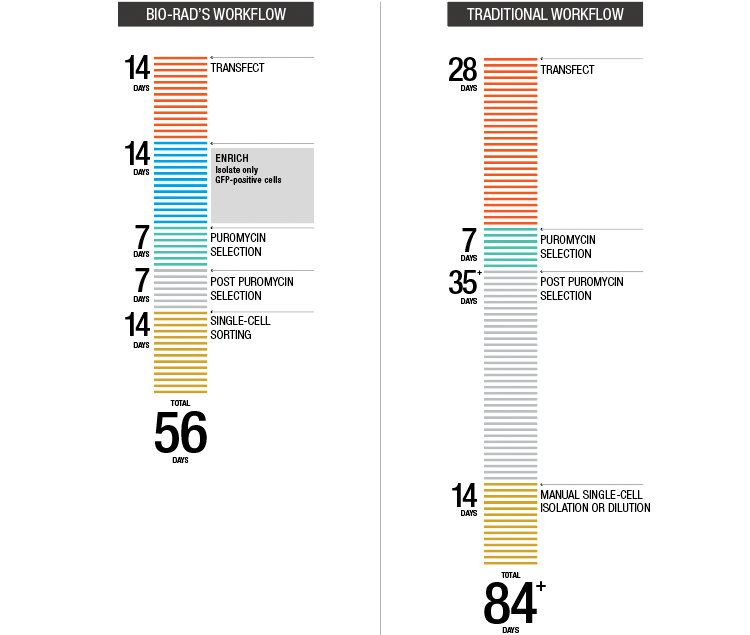
Incorporating the S3e Cell Sorter can shorten gene editing workflows by about 30 days.
Researchers used an S3e Cell Sorter in a core lab at Columbia University Irving Medical Center for this study. Hear the core lab managers’ perspectives on the S3e Cell Sorter.
The S3e Cell Sorter works quickly, enabling high-throughput screens, like the one performed in this study, which included 204 different ORFs. Despite its speed, the S3e Cell Sorter is gentle. Preserving cell viability is critical for most gene editing applications, which typically require live, healthy cells for downstream analysis.
The S3e Cell Sorter was designed to be easy to use, even for researchers unfamiliar with flow cytometry. Its intuitive interface and simple operation make integrating cell sorting into a gene editing workflow painless. Unlike many cell sorters, the S3e Cell Sorter features walk-away operation, so there is no need to continuously monitor the instrument during a sort.
References
Nambiar T et al. (2019). Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nature Comm 10:3395.

