The polymerase chain reaction (PCR), developed by Kary Mullis and colleagues at the Cetus Corporation, has had a profound impact on life science research. Following its publication in 1985, scientists could select an exact DNA sequence and copy it millions of times over with a straightforward, fast procedure. How would they apply this powerful new tool?
One scientist who grappled with this question was Alec Morley. At the time the head of the hematology department at Flinder’s University in Adelaide, Australia, Morley was on his way home from a conference in Sydney in 1987 when the potential of PCR struck him. “I’ve had a couple of ‘Eureka!’ moments in my life, but this was a particularly good one,” he remembers. “I was sitting on a plane taxiing out of Sydney Airport, and the question suddenly came to me: what specific gene sequences in the leukemias could be detectable by PCR?”
Learn about the development of an early digital PCR method by Alec Morley and his colleagues at Flinders University in Adelaide, South Australia, and their use of Droplet Digital PCR technology today.

Department of Haematology
Flinders University

Researcher Sue Latham begins the Droplet Digital PCR workflow by using the droplet generator to partition her sample into approximately 20,000 droplets per well.

Sue Latham and Alec Morley examine ddPCR™ results from the droplet reader, which analyzes each droplet for positive or negative florescence. When the analysis is complete, the results provide an exact concentration of target DNA in the sample.
How could Morley and his colleagues apply PCR to leukemia detection? Answering this question would yield important findings about that illness. Yet even as Morley’s research group searched for answers using PCR, they were also innovating with the technique. Today, Morley and his colleagues Pam Sykes, Sim Neoh, and Mike Brisco are recognized as having pioneered the concepts behind digital PCR, a significant refinement of the original PCR method and now a promising research technique in its own right (Baker, 2012).
PCR, the amplification of a chosen DNA fragment, becomes “digital” through sample partitioning. In other words, instead of the amplification occurring in one tube, a digital PCR spreads the experimental sample and the amplification across hundreds or even thousands of tubes. This allows for binary results — target DNA is either present in one or more of the multiple tubes, or it isn’t. Applying statistics to the group and its distribution of positives vs. negatives produces a meaningful numerical result. The more partitioned the sample is, the greater statistical precision the digital PCR has. This new level of sensitivity holds huge potential for precisely measuring genetic events such as cancer mutations, genomic variations, and gene expression. It’s implementing this technique that has, so far, proven most difficult.
Indeed, after bringing together the concepts behind digital PCR and successfully using a basic system, Morley’s group ultimately gave up their technique as too labor intensive. However, almost twenty years later, they would find a viable new sample partitioning technology: Bio-Rad’s Droplet Digital PCR system. Using advanced microfluidics, this system “digitizes” the PCR reaction by spreading it across 20,000 nanoliter-sized droplets, creating the high data resolution needed for counting sequences with precision. Today, Morley’s group uses a ddPCR system to addres a new challenge in measuring leukemia, thus bringing their work with digital PCR full circle.
“It’s a bit of a buzz that we’re using Droplet Digital PCR, a technique which we pioneered and then gave up for many years,” laughs Morley. “Our contribution, for what it was worth, was unknown until the last year or two. That it has reemerged is very personally pleasing to me, to Pam Sykes, to Mike Brisco, Sim Neoh — all the members of the team.”
Two Strands of Research
Alec Morley decided to apply PCR to leukemia detection in 1987. The conventional way of assessing leukemia at that time, Morley explains, was by looking through a microscope at a bone marrow sample. When a patient presents with, say, acute lymphocytic leukemia, it’s easy for a trained pathologist or hematologist to spot the many leukemic cells. However, Morley specifies, “once the proportion of leukemic cells drops below that 1%, 1 to 2%, you can’t detect them by eye. The patient might be cured, they might not. It’s a black box; we don’t know. So, to follow the effects of treatment, and to possibly adjust treatment, you need more sensitive detection.” Opening the “black box” would help doctors spot potential relapses—a major problem in treating the illness—while also sparing people who didn’t need them from the intensities and side effects of additional treatments.
As he realized the potential of PCR during his “Eureka!” moment, Morley also considered possible targets in leukemia for the gene amplification technique. “There were three,” he recalls. “The ras proto-oncogene, the BCR-ABL translocation in chronic myeloid leukemia, and immunoglobulin T-cell gene rearrangements in the acute leukemias.”
Mike Brisco, a young molecular biologist who joined Morley’s group around that time, became instrumental in making immunoglobulin and T-cell gene rearrangements the most favorable of the three PCR targets to investigate first. “What we were looking at with these gene rearrangements was the mechanism the immune system uses to generate diversity, so it was a huge range of genes,” says Brisco, who has continued to investigate PCR methods at Flinders ever since.
Because of this immune mechanism, each patient presenting with acute lymphoblastic leukemia (ALL) created a unique clone in their leukemic B or T cells. “Nailing down a usable sequence was quite a tricky problem to solve. We were lucky,” recounts Brisco, “in that we found parts of the sequence upstream and downstream of the unknown rearrangement that were well-conserved, so we could design primers for them, then amplify up the bit in between.” With Brisco’s work, which took a year, the group now had a way to get patient-specific markers for ALL.
The research that yielded his group’s digital PCR method was like “two strands coming together,” says Morley. Isolating the gene rearrangement gave them one strand. Morley and his colleague, geneticist Pam Sykes, focused on the intellectual effort of assembling the method. Morley elaborates, “Having been able to detect gene rearrangements by PCR, the next question was, can we quantitate this? Because if you can actually measure things, that’s much more powerful than just detecting them.”
To answer that question, Morley and Sykes brought in the second strand of research by adapting methods from the cell culture work in the laboratory. Morley had developed a technique for cloning lymphocytes to study human somatic mutations. By distributing the growing cells among multiple wells, he found that the growth of the mutant cultures followed the Poisson distribution, a distribution pattern used to describe the probability of the occurrence of random events. He eventually developed a quantification method. “We took that quantification method from lymphocyte cloning and combined it with the markers that [Brisco] had developed,” Morley says. “So instead of cloning a cell in a well, we cloned a molecule in a tube with a PCR.”
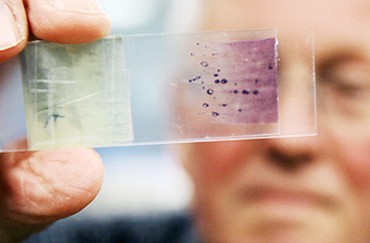

Left: Alec Morley displays a slide of a bone marrow section. A trained hematologist or pathologist can detect leukemic cells at a concentration as low as 1% under a microscope. Right: Monica Dreimanis checks the growth of mutant B-cell lines. In developing their early digital PCR system, Morley and Pam Sykes applied a quantification method based on the Poisson distribution pattern which Morley had previously used to quantify cell cultures, to PCR in order to obtain a quantitative result.
Putting Digital PCR into Practice
In their effort to apply PCR to leukemia detection and measurement, Morley’s group had scoped out a marker for ALL. They had a quantification method they had adapted from cell culture. Now they had to combine them to put this system into practice. That task fell largely to Simhee Neoh, another young researcher who had recently joined Morley’s group.
“It was a fairly laborious thing to do umpteen manual dilutions of samples [to find a point where both positives and negatives would fit within a dynamic range],” Neoh remembers, “and then check every single interaction between the leukemic primers.” Once the right dilution was found, Neoh then had to perform the multiple PCR reactions, run a gel for each, and photograph the results with a large-format Polaroid camera. Only when numerous gels had been run could they aggregate the results, perform the Poisson analysis, and arrive at a quantification.
Contamination also posed a major concern, given the many different samples needed and nested PCR reactions being performed. “We set up a clean room at the other end of the building, so there were two discrete labs,” Neoh says. “We had separate lab coats, separate everything because of the fear of contamination.”
A Very Sensitive, Very Impractical Technique

Mike Brisco played a crucial part in targeting the immunoglobulin T-cell and B-cell gene rearrangements that served as target sequences for the group’s work in developing an early digital PCR system.
Their digital PCR process, which they referred to as “limiting dilution PCR,” proved complex and time-consuming. Nonetheless, the group’s careful work paid off. Using an archival set of samples, “we were able to measure acute leukemia levels, which varied quite a lot during treatment,” says Brisco. “We were quite pleased as well to be able to detect levels lower than [those detected with] microscopes. Instead of 1%, we were able to get as low as spotting, say, 0.01% leukemic clones.”
“We were really blown away by how sensitive the technique was,” Brisco continues. “It wasn’t just slightly more sensitive than the other techniques that were being used, it was a lot more sensitive.”
This dramatically improved sensitivity enabled them to open up the “black box” of minimal residual disease in ALL, a limit which until then had blocked understanding of disease progression. Morley describes how, using their digital PCR process, “we were able to divide patients into three groups depending on their level of minimal residual disease: those who are cured, those who relapse, and a middle group. That division has been confirmed by many much larger studies since and has become the basis for modifying treatment.”
The group described these findings in several publications, culminating in a 1994 study in The Lancet (Brisco et al. 1994). “They were exciting times,” remembers Morley. “At the American Society of Hematology [meeting], the person giving the session said, ‘If this is confirmed it’s going to change the way we treat leukemia.’ And it was confirmed.”
As they were publishing their leukemia studies, Morley and his colleagues also published their digital PCR work in Biotechniques as a general quantification method (Sykes et al. 1992). This paper is often cited now as one of the key early descriptions of digital PCR methods. Yet even as they had success with their approach, Morley’s group began to move away from it and toward real-time PCR. “It was a good method,” Morley says. “But it wasn’t practical. We didn’t have the technology in those days; all we had was two pairs of hands.”
Answering a New Question with Droplet Digital PCR

Alec Morley, Brad Budgen, and Sue Latham discuss data from the Droplet Digital PCR system. They are working with Monoquant, a company started by Morley to commercialize intellectual property associated with his research, to develop a diagnostic test to monitor chronic myeloid leukemia using Bio-Rad’s ddPCR technology.
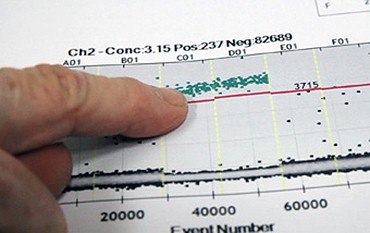
Morley and his colleagues are applying ddPCR to get a more precise quantification of the BCR-ABL translocation, the underlying cause of CML, by analyzing patient DNA rather than RNA, as is current practice.
Today, Morley continues his effort to find better ways to measure and monitor leukemia. For several years he has been working through his company, Monoquant, to develop a technique for quantifying chronic myeloid leukemia (CML) using a patient’s DNA. This is a departure from the current method, which is to analyze RNA expression using real-time PCR.
To streamline his DNA-based approach, Morley turned to Bio-Rad’s ddPCR technology as a way to get a highly precise measurement of CML’s signature BCR-ABL translocation. “Droplet Digital PCR essentially does what Sim and Mike used to do manually,” says Morley. “Each droplet contains a PCR reaction that is read for positive fluorescence or negative fluorescence. Instead of 25 laboriously set up electrophoresis gels, you get 20,000 droplets set up in a few minutes and read in a short time.”
This massive number of droplets creates the data resolution needed for absolute quantification of target sequences and doesn’t require a standard curve, an advantage over the relative quantification obtained through real-time PCR. “We believe Droplet Digital PCR may enable us to provide a more precise quantification of the BCR-ABL translocation,” Morley says.
While quantifying BCR-ABL at the DNA level allows for more sensitive detection, it also requires a patient-specific approach. Each translocation occurs at a unique breakpoint in the genome, yet these differences get spliced out in the RNA transcription process. To get the specificity necessary to make their results useful, Morley created a system of primers in developing his assay. “You have different primers for different patients, and they may vary in efficiency,” Morley elaborates. “Droplet Digital PCR is less affected by variations in amplification efficiency using these various primers. We think this may make regulatory bodies look more favorably on our using a DNA target.”
Morley is conscious of practicality as well as precision, given both the needs of his lab and that he wants to develop a diagnostic test for wide application. “In choosing between different instruments, one really needs to look at what one’s specific needs are,” he says. “It’s really what we would call a ‘horses for courses’ thing. The Bio-Rad [ddPCR technology] does what we want, at the level of sensitivity that we need, and it’s quick. So it was the obvious choice.”
“Measurement Is a Great Thing”
“I got into hematology because it’s an area where things are quantifiable. You can answer questions because you can measure things,” Morley reflects. “That’s a great thing. Measurement is a great thing.” Indeed, Morley’s career has centered on what he calls “the needle in the haystack problem,” the need to pinpoint and quantify an incredibly small—yet highly significant—marker that will help people with illnesses.
At the core of this pursuit is Morley’s belief that if he can just ask the right question, he can find a measurable answer. “Einstein said, ‘God doesn’t play dice with us,’” Morley states. “The answers are all there. It’s up to us to formulate questions. That’s why I like technology; because it helps you answer something. Formulate the question, and then go and get the piece of technology that will enable you to answer it.”
From innovating with PCR in its early stages to applying Droplet Digital PCR technology today, Alec Morley has done just that.
References
Baker. Digital PCR Hits Its Stride. Nature Methods 9, 541-544.
Brisco MJ et al. Outcome prediction in childhood acute lymphoblastic leukaemia by molecular quantification of residual disease at the end of induction. The Lancet 343, 196–200.
Sykes P et al. Quantitation of targets for PCR by use of limiting dilution. BioTechniques 13, 444–449.

