When you hear the words cellular computation or cellular memory, you probably think of instant messaging, navigating, internet browsing, video streaming, or even near field communication (NFC) using the sophisticated cell phone in your pocket. But researchers have begun exploring an extraordinary alternative cell computational system, the cells that make up our bodies.
The Origins of Computing
The word computer originally referred to one who computed, a human being who crunched numbers. To assist with such number crunching, the British pioneer mathematician Alan Turing in 1936 conceived of a machine, the Turing machine, that could process and solve any computable problem.
The idea of the Turing machine is very straightforward. Simply imagine you are the machine. You are holding a pencil and an infinitely long piece of paper. On this piece of paper is a single line of symbols, which you are instructed to read one at a time. You also have instructions to write and rewrite symbols based on a specific set of rules. Once you are done with one cycle of read/write, you move on to the next symbol, and keep going. This is exactly what the Turing machine does. Although the machine’s design is simple, the logic behind it is able to simulate any modern computer algorithm and the concept of Turing’s machine lives on as the basic building block at the heart of all modern computers — the central processing unit, or CPU.
Cells in Our Body Are Not so Different From the Cell Phones in our Pockets
Living organisms carry out very complex physical processes, which are dictated by many complex and sophisticated entities: cells. Within each cell, layer by layer, elementary components and biochemical operations are ruled by instructions stored in the cell’s genome, encoded by sequences of nucleic acids, or DNA. Strikingly, living cells, biomolecular machines that process DNA and RNA, do work very similar to that of the Turing machine. DNA is transcribed into RNA, ribosomes then read information encoded by the RNA and translate it into the polypeptide chains that form proteins. Proteins then regulate and dictate which segment of DNA is to be transcribed into RNA and the cycle continues (Figure 1). Despite emerging from very different domains, the inner workings of biomolecular systems and the Turing machine are quite similar and the two share a computational philosophy.
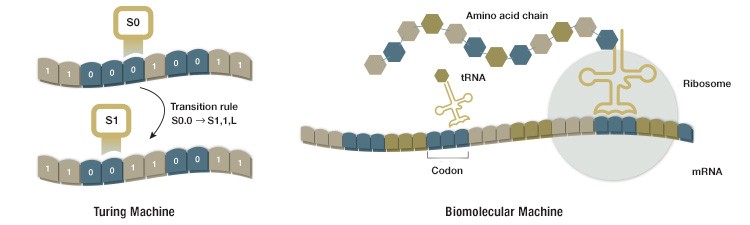
Fig. 1. The Turing machine and a ribosome share many features. Both systems read information stored as strings of symbols and process these symbols according to a defined set of rules. The Turing machine writes and rewrites symbols, one at a time, on a strip of paper according to specific transition rules. The ribosome reads mRNA sequences and adds amino acids to a growing amino acid chain according to rules defined by the genetic code.
Turning Cells into Computers
In fact, the similarities between the man-made Turing machine and the biomolecular machines of living cells are so striking that they have captured the attention of mathematicians and physicians. In 1982, Charles Bennett, from IBM’s Watson Research Center, propounded the hypothetical concept of a molecular Turing machine and speculated that biomolecules might one day become the basis of more energy-efficient computers. However, it was not until 1994, when University of Southern California professor Leonard Adleman solved the Hamiltonian path problem using DNA, that the concept of DNA computing (or cell computation) was first demonstrated to be feasible in the real world (Adleman 1994). Adleman’s findings generated enormous scientific and public interest, which kick-started cell computation research and development. However, building a biomolecular computer was not as easy as expected. Many technical challenges arose and such aspects as programmability, logical circuit design, and autonomous computing still await complete solutions.
The Bacterial Cell as a Recording Device
One of the critical problems in designing sophisticated synthetic biomolecular circuits is how to design autonomous cellular memory – the ability to convert information from one format into another according to a defined logical procedure and then store this transient information in a long-lasting and scalable biological database. In 2014, Farzadfard and Lu of the Massachusetts Institute of Technology proposed and demonstrated a programmable and modular cellular architecture called Synthetic Cellular Recorders Integrating Biological Events, or SCRIBE, to address critical questions about autonomous cellular memory.
SCRIBE is a scalable platform that converts the genomic DNA of living cells into “tape” for memorizing information. In addition, it allows recording, rewriting, and reporting of analog information, such as magnitude and time span of exposure, by altering the sequence of nucleic acids. Confused? Let’s think of a living cell, such as bacterial cell. If we consider the exposure to light or a chemical of a bacterial cell as a recordable voice, then the genomic DNA can be imagined as a tape on which to store such recordings with the cell as the recorder. Now let’s break down this recorder into its individual components.
Typically, a recorder carries out the following basic functions:
| Record | Converting a voice from one data format (analog) to another data format (digital) |
| Write/Rewrite | Writing or erasing and rewriting the information on a tape |
| Data storage | Transferring information to a medium (tape) on which it is stored permanently |
| Read | Decoding information stored on a tape |
| Play | Conversion of stored information from one format (digital) to another (sound) |
Bacteria already carry out many of these functions. In bacterial systems, exposure to the chemical IPTG activates lac promotor activity and results in protein expression. In this case, the bacterial cell senses extracellular analog information in one format (chemical concentration of IPTG) and converts it into another format (protein expression) inside the cell. Therefore, the concept of “record” in bacterial cells can be understood as receptor sensing and signal transduction.
To build a bacterial computer, a way must be found to permanently store transient information. Once an extracellular signal or message is sensed and converted from one format to another, a medium is needed to store this message. Because of its high storage capacity, durability, fidelity, and ease of replication, genomic DNA, the basis of heredity that is passed from one generation to the next, is actually an ideal storage medium to support not only life but also DNA computing. “Data storage,” then, would be permanent changes to the genomic DNA sequence.
Nowadays, there are a variety of techniques to “read” or “play” genomically encoded information in a living cell. For example, gene expression profiling and functional reporter assays, such as promoter-driven expression of GFP, are standard ways to play genetic information, while high-throughput DNA sequencing allows us to read large quantities of genetic information. Therefore, “read” and “play” functions in living cells can be easily achieved using genomic DNA as a storage medium.
If the record, data storage, read, and play functions can be mimicked by different biochemical events and biomolecules in a living cell, the biggest remaining hurdle of autonomous cellular memory would be the “write/rewrite” function, the process of capturing and converting transient messages into sequences of nucleic acids that can be stored in the genome. It was previously demonstrated that synthetic oligonucleotides (single-stranded DNA) that overexpress β-recombinase of bacteriophage λ can be efficiently and specifically recombined into target genomic loci through homologous recombination when delivered into bacterial cells by electroporation (Yu et al. 2000). This result demonstrated the feasibility of writing specific messages onto a defined location of genomic DNA. However, this approach cannot be converted into an autonomous recording system because it requires exogenous delivery of oligonucleotides and the presence of these oligonucleotides is not a direct result of the extracellular signals to be read and recorded.
SCRIBE Paves the Way to an Autonomous Cellular Recording Device
In order to solve the writing problem, Farzadfard and Lu developed SCRIBE, a modular system that leverages the bacterial retron-msDNA system to autonomously generate unique single-stranded DNA intracellularly in response to a defined external signal. The retron-msDNA cassette encodes three components — a reverse transcriptase (retron RTase) and two RNA moieties (msr and msd) — in a single transcript. The msr–msd RNA, once transcribed, folds into a secondary structure, guided by base pairing. The retron RTase encoded by the retron transcript recognizes this secondary structure and uses a conserved guanosine residue in msr as a priming site to reverse-transcribe the msd sequence. This process produces a unique single-stranded hybrid RNA-DNA molecule called a multicopy single-stranded DNA (msDNA) (Lampson et al. 2003). Because msDNA can be carefully engineered to carry specific sequences, precise messages can be incorporated into target genomic loci of living cells by co-expression with bacteriophage β-recombinase.
The generation of message-carrying msDNA and the expression of β-recombinase are both regulated by a bacterial expression system that is tied to the sensing system; therefore, their presence is precisely controlled in response to a defined extracellular signal. Interestingly, because intracellular expression levels of msDNA and recombinase are proportional to extracellular stimulation, the magnitude and duration of extracellular signal exposure can be recorded as an increase of recombination rate at specific loci within a living cell. In addition, integrating several interdependent retron-msDNA systems allows construction of logic gates, such as AND, OR, and NOR gates, that enable the design of more complex operational circuits in living cells.
Using SCRIBE to Build Next-Generation Computers
SCRIBE is a simple yet elegant proof-of-concept system for autonomous cellular memory. It proves the feasibility of long-term autonomous cellular recording and memory functions in response to environmental changes in living organisms, and Farzadfard and Lu provide several examples that demonstrate its potential applications in biological and biochemical systems engineering. Although the current model has yet to be adapted for real-world applications, the conceptual work that Farzadfard and Lu demonstrate is a valuable step forward in cell computation. Hopefully, the application of the SCRIBE platform will open up a broad range of new capabilities for engineering biology. One can imagine, in the foreseeable future, general-purpose computers that consist of nothing more than a single living cell, and applications for these nifty DNA computers that range from monitoring disease to targeting the release of drugs inside our bodies.
References
Adleman LM (1994). Molecular computation of solutions to combinatorial problems. Science 266, 1,021–1,024.
Lampson BC et al. (2003). Retrons, msDNA, and the bacterial genome. Cytogenet Genome Res 110, 491–499.
Yu D et al. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97, 5,978–5,983.

