
Senior Scientist
Sangamo BioSciences, Inc
Timothy Ray Brown is the only person who has been cured of HIV to date (Pollack 2011). Dubbed the Berlin Patient, the now 46-year-old Brown was living with HIV and leukemia in Berlin, Germany when he received two bone marrow stem cell transplants from a donor who carried, in both copies of his CCR5 gene, a well-characterized natural defect, the CCR5-∆ 32 mutation. (CCR5 is a major co-receptor used by HIV to enter and infect CD4+ cells.) The CCR5-∆ 32 mutation rendered the CCR5 protein inactive and thus made the donor’s cells naturally resistant to HIV. After completing the transplants in 2007 and 2008, Brown immediately stopped taking antiviral drugs and has remained HIV-free ever since, igniting considerable interest within the cure research community.
Sangamo BioSciences, Inc., located in Richmond, California, is using its proprietary zinc finger DNA-binding protein (ZFP) technology to develop therapies designed to provide a functional cure for HIV. ZFPs are a class of naturally-occurring proteins that can be engineered to recognize and bind to defined DNA sequences. By engineering gene-editing ZFP nucleases (ZFNs), it is possible to permanently disrupt CCR5 in immune cells (CD4+ T cells), thereby making them resistant to infection and available, potentially to control the virus without drugs.
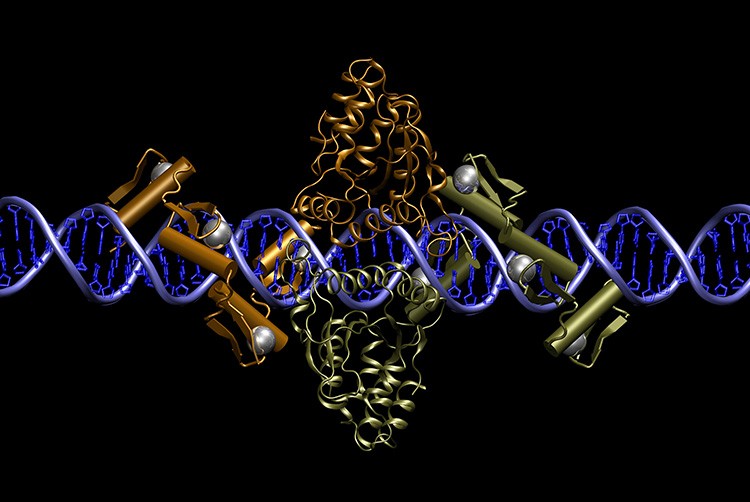
Pair of zinc finger nucleases bound to DNA. Courtesy of Dr Jeff Miller, Sangamo BioSciences, Inc.
“Sangamo’s technology is unique in that it allows you to work at the DNA level to modify a gene at the core of a disease,” says Dr. Gary Lee, senior scientist at Sangamo. “To combine the zinc finger gene editing technology with what we learned from the delta 32 mutation in nature, and then produce an effective therapeutic, is really exciting.”
From top to bottom: Gary Lee and intern John Campo exploring protein structure using PyMOL; Sal Orlando, research associate, examining precipitates and loading the QX100 reader component; Travis Wood, development manager, Gary Lee, and Joshua Kahn, senior research associate, in front of Sangamo BioSciences, Inc.
Low Copy Event Detection
As HIV infection progresses, the number of functional CD4+ T cells drops, eventually leading to acquired immunodeficiency syndrome (AIDS), the symptomatic stage of infection. Because HIV DNA integration in the host cell’s genome is a rare event, occurring in only 1/1000 to 1/10,000 of CD4+ T cells, one of the major challenges in Sangamo’s research is measuring the level of HIV DNA in a subject to evaluate the therapy’s effectiveness.
Initially Sangamo used real-time PCR (with TaqMan hydrolysis probes) to detect and quantify HIV DNA in subjects’ samples. However, real-time PCR proved inefficient for low copy event detection. In such cases, excessive background genomic DNA (gDNA) competes against target DNA (HIV DNA) in each PCR. To capture target DNA, gDNA had to be diluted several fold, and the target still could not be detected with accuracy.
It became obvious to Sangamo’s scientific team that a more sensitive and precise method was needed to detect and quantify HIV DNA in blood and other lymphoid samples with the reliability required for clinical studies. The novel Droplet Digital PCR (ddPCR™) technology, which can detect single copies of a target sequence within a complex sample, provided the answer.
“With real-time PCR, we couldn’t tell whether our data were a result of experimental variation or biologically real. At low copy numbers in two separate assays, we would see a tenfold difference,” says Lee. “With the ddPCR technology, the difference was only twofold, which is data we can really trust.”
Droplet Digital PCR
The QX100™ Droplet Digital™ PCR system consists of two components, the droplet generator and the droplet reader, which are used in conjunction with a regular thermal cycler. The droplet generator uses micro fluidics to partition each sample into 20,000 water-in-oil nanoliter droplets. Following PCR, each droplet can be assigned as positive or negative based on fluorescence, which corresponds to the presence of the target DNA sequence. Each nanodroplet serves as an independent amplification event where reaction specificity is increased due to dilution of background DNA (Figure 1), enabling resolution of rare target DNA sequences (Hindson 2011).
PCR amplification is conducted in the presence of TaqMan hydrolysis probes, allowing detection of positive droplets based on their fluorescence signals using the droplet reader. As the fluorescence signal of the PCR is converted from analog to digital, there is no requirement for a standard curve. Positive and negative droplets are used to calculate the concentration of target and reference DNA sequences by exploiting Poisson sampling statistics. Sangamo was one of the early adopters of the ddPCR technology, acquiring the instrument in the summer of 2011.
“Since acquiring the QX100, we’ve been a lot more confident in what the assays are telling us. We were amazed at how easily, accurately, and reproducibly we could measure low copy events in a sample of genomic DNA,” says Lee. “We’re able to generate data that is a lot more accurate for our research.”
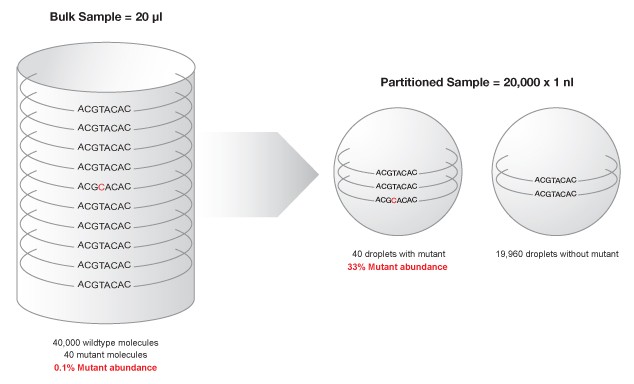
Figure 1. Partitioning minimizes competition between target sequence and highly abundant background DNA, enhancing mutant detection.
Sangamo’s New Clinical Trials
Sangamo’s HIV therapeutic is a cell therapy that uses an individual’s own T cells, which are modified with ZFNs, cultured, expanded outside the body, and then reintroduced into the donor. Sangamo develops the modified resistant CD4+ T cells over a ten-day period. The CCR5-knockout cells are then subject to testing to ensure quality standards. Once the product is released, the cells are re-infused into the patient.
Currently, Sangamo is conducting two new phase 2 clinical trials in HIV-infected subjects. The trials are designed to increase the number of engrafted T cells in which CCR5 is biallelically modified (modified in both copies of the gene) by ZFNs. The design is based on the success of a recent phase 1 study in which a treated subject who was heterozygous for the CCR5-∆ 32 mutation was observed to have undetectable HIV in his blood during a treatment interruption (TI), a period during which he stopped taking the conventional antiviral therapy known as HAART (Pollack 2011). As a heterozygote undergoing the ZFN-mediated treatment, the patient – called the Newton Patient — ended up with roughly double the number of biallelically modified T cells. Data from that study, combined with Sangamo’s Phase 1 study, demonstrate a statistically significant relationship between the number of biallelically-modified engrafted cells and a reduction in viral load during TI. Sangamo’s trial is testing the approach in a larger group of CCR5-delta 32 heterozygotes.
The company is conducting a second phase 2 clinical trial to enhance modified cell engraftment. In this protocol, subjects undergo a brief preconditioning treatment of cyclophosphamide (Cytoxan), which transiently depletes lymphocytes (including T cells) in the body. By reducing host T cells, the ZFN-modified T cells will better engraft within the body. The effectiveness of the therapy in reducing HIV levels is monitored over a three-month treatment period through blood screening assays that include the use of the QX100™ ddPCR™ system.
“The QX100 system is really a godsend for us, as we can now more accurately detect the HIV DNA level — the HIV reservoir — in these subjects,” says Lee. “We are much more confident in determining the HIV reservoir level than ever before.” For Sangamo, the ability to employ ddPCR technology to accurately detect HIV reservoir levels will be instrumental in the development of their functional cure therapy.
References
Sangamo’s HIV therapy studies demonstrate how ddPCR technology can be applied for rare event detection applications in clinical studies. The QX100 system plays a central role in the company’s research, providing highly precise, reproducible data in support of functional cure development.
Reference
Hindson BJ et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83, 8604–8610.
Perez EE et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26, 808–816.
Pollack A (2011). New Hope for a Cure for H.I.V. The New York Times. Accessed June 12, 2012.

