RNA sequencing (RNA-Seq) is a fundamental discovery technique for determining the expression patterns of numerous encoded genes in a biological process or disease state through unbiased detection of both known and unknown targets. RNA-Seq has led to innumerable breakthroughs across life sciences and medicine; however, a need remains for new sequencing and library preparation technologies to address gaps in current methods. This article discusses how improved access to RNA-Seq benefits translational research and summarizes how a novel RNA library preparation chemistry can enable the acquisition of robust data from new sequencing platforms, with suggestions for further areas of innovation.
New Short-Read Sequencers Promise Increased Access to RNA-Seq
The contest to disrupt the next-generation sequencing market has begun, as new short-read sequencing platforms aim to offer sequencing that is rapid, accurate, and more affordable than ever before. For RNA-Seq, researchers can now choose from a variety of excellent platforms.
Common applications for next-generation sequencing include exome, transcriptome, whole genome, and targeted sequencing. However, the scope of what can be done with RNA-Seq is limited — application to certain organisms and sample types, such as fixed tissue samples, remains challenging.
Improving the accessibility of RNA-Seq has the potential to produce positive ripple effects across both basic and translational fields of research. In basic research, broader adoption could improve genome annotations and increase understanding of the functions of many recently discovered genes (Mu et al. 2020). Findings from RNA-Seq also show unique clinical utility, specifically in detecting biomarkers and profiling tumor protein expression in formalin-fixed paraffin-embedded (FFPE) samples, which are arguably the most common archived biospecimen available. In a recent study, detection of brain tumor gene fusions in FFPE samples by next-generation mRNA sequencing led to decisive diagnostic relevance in 43% of cases, while in 40% of the cases, the detection of fusion genes provided a druggable target for those patients (Stichel et al. 2019). Given limitations in the diagnosis and treatment of brain tumors, such outcomes are incredibly significant. Such advances are not limited to brain tumors — for example, RNA-Seq clinically applied to breast cancer enables the prediction of prognosis and other outcomes (Chen et al. 2022, Xia et al. 2022).
Enabling more discoveries like these requires the introduction of new sequencing platforms and technologies that make RNA-Seq available to a broader number of researchers. Moreover, with improved accessibility, sequencing platforms could also speed development and empower new clinical sequencing applications in predictive diagnostics.
The Role of RNA Sequencing Library Preparation in Technology Disruption
Ensuring that new short-read sequencing platforms reach their fullest potential goes beyond the instrument purchase price, RNA isolation, and sequencing reagents — it also requires library preparation reagents that can enable high-quality sequencing outcomes for a broad range of sample conditions.
Library preparation is a key step in RNA sequencing and involves the conversion of RNA to cDNA. An adapter ligation step introduces a sequence to the 3’ end of the RNA transcript for use by the reverse transcriptase during first-strand cDNA synthesis. Subsequently, amplification, size selection of specific fragments, and cleanup occurs, after which the cDNA library can be run on the sequencer. Following the sequencing run, read lengths of 50–300 base pairs (bp) are analyzed separately and assembled into a whole genome sequence using open-access scripts during bioinformatic analysis. To identify the transcripts detected, the analysis compares the data pipeline to a reference genome for that species.
RNA sequencing needs to be suitable for a range of sample types to ensure broad adoption. While many library preparation options are available for fresh samples and isolated cell cultures; fixed tissues, such as FFPE samples, remain a challenge for the high RNA quality requirements of many commercial RNA-Seq library prep kits. The RNA isolated from FFPE samples tends to be of poor quality, as it is often heavily degraded and chemically modified from crosslinking during fixation. The ability to use RNA from fixed tissues for sequencing would allow greater access to sequencing data from tumor biopsies, which are typically stored in FFPE formats, and remain the most common archived specimen type. Thus, developing protocols for FFPE samples would make numerous sample archives available for valuable clinical research.
Enter the SEQuoia family of RNA library preparation kits. The SEQuoia Complete Stranded RNA Library Prep Kit not only works with a variety of sample types but can also provide complete transcriptome profiling of both long and small RNAs, in contrast to other methods that capture a specific size range. For research where speed and ease are key considerations, the SEQuoia Express Stranded RNA Library Prep Kit offers a rapid 3-hour, 3-tube workflow that detects long RNAs from higher-quality RNA samples.
SEQuoia Kits Enable Whole-Transcriptome Analysis of FFPE Samples
To address the challenges of RNA library preparation from FFPE samples, a recent study evaluated the SEQuoia Complete Kit for the capture of the whole transcriptome from FFPE samples, including both long and short RNA biotypes, in a breast cancer study (Perike et al. 2022). The SEQuoia workflow used the SEQuoia Complete Kit followed by a novel post-library method to remove the ribosomal RNA-derived cDNA fragments with the SEQuoia RiboDepletion Kit. This workflow was compared to a popular commercial kit that uses a standard pre-library ribodepletion strategy.
The SEQuoia Complete and SEQuoia Express Kits eliminate common challenges associated with RNA-Seq by first eliminating bias introduced by typical multistep enzymatic processes. Using a single unique engineered retrotransposon, SEQzyme, in place of the traditional reverse transcriptase and ligases found in other RNA-Seq library preparation kits produces cDNA libraries that more closely resemble the source transcriptome. SEQzyme provides better processivity and enables end-to-end template jumping using a continuous synthesis reaction that converts RNA to cDNA and adds sequencing adapters in a single step. In addition, in the SEQuoia Complete Kit format, the RNA fragments are first polyadenylated and then primed with a polythymidine sequence on the 3′ end of the sequencing adapter. As a result, both long (>200 bp) and short (<200 bp) RNA transcripts are uniquely captured in a single library workflow.
A second advantage of the SEQuoia workflow is that utilizing a novel post-library ribodepletion strategy better preserves the richness of the transcriptome and avoids the loss of small RNAs by depleting ribosomal RNA after its conversion to cDNA — a point when the sample is more stable — and not using size-selection at that stage. These unique attributes ensure a more robust process, leading to more complete transcriptome profiling.
In the study of breast cancer samples, RNA-Seq library preparation using the SEQuoia workflow showed the ability to provide both short and long RNAs from a single workflow, overcoming the biases inherent to the other commercial kit, which only captured long RNAs (Perike et al. 2022).
This work shows the promise of RNA sequencing to unlock novel molecular mechanisms so that future, biologically-driven studies can occur even with FFPE and other degraded or low-yield RNA samples.
Excellent Library Preparation Is Key to RNA-Seq Performance
Recent studies demonstrate that SEQuoia Kits can be easily adapted for compatibility with new short-read sequencing platforms. As shown at the recent Advances in Genome Biology and Technology (AGBT) 2023 conference, the SEQuoia Complete and SEQuoia Express Kits excel not only on a standard sequencer, but can also offer a rich data set on new short-read sequencer platforms such as the Element Biosciences AVITI System (Bio-Rad bulletin 3498) and the Singular Genomics G4 Platform (Kumar et al. 2023).
As summarized in Table 1, the SEQuoia Complete Kit can enable the capture of a complete transcriptomic profile, including both long and short RNA transcripts, in a single workflow, and the resulting libraries can be analyzed on the Element AVITI System as well as the Singular G4 Sequencing Platform (Kumar et al. 2023; Bio-Rad bulletin 3498). The SEQuoia Express Kit offers a rapid 3-hour, 3-tube workflow that detects long RNAs from higher-quality RNA samples (Table 1) and is also compatible with the Element AVITI System and Singular G4 Sequencing Platform (Kumar et al. 2023; Bio-Rad bulletin 3498).
Table 1. Comparison of SEQuoia RNA-Seq Library Prep Kits.
| SEQuoia Complete Stranded RNA Library Prep Kit | SEQuoia Express Stranded RNA Library Prep Kit | |
| Capture range | All RNA subtypes, >20 bp | Long RNA fragments, >200 bp |
| Input range | 100 pg–1 µg | 1 ng–1 µg |
| Sample type | All sample types, including low-quality formalin-fixed paraffin-embedded (FFPE) samples | Standard good-quality sample |
| Sequencing read | Single read | Paired end |
| Protocol time | 4 hr | 3 hr |
| RNA subtypes captured | mRNA, long noncoding RNA (lncRNA), miRNA, snoRNA tRNA, and more | mRNA, lncRNA |
| Tubes per kit | 7 | 3 |
To highlight the data quality, SEQuoia Complete libraries run on the Element AVITI System produced consistent sequencing results in long RNA transcript detection among sample replicates and different RNA input levels (Figure 1A) and also detected small RNA transcripts with high (100 ng) or low (10 ng) RNA input (Figure 1B) (Bio-Rad bulletin 3498).
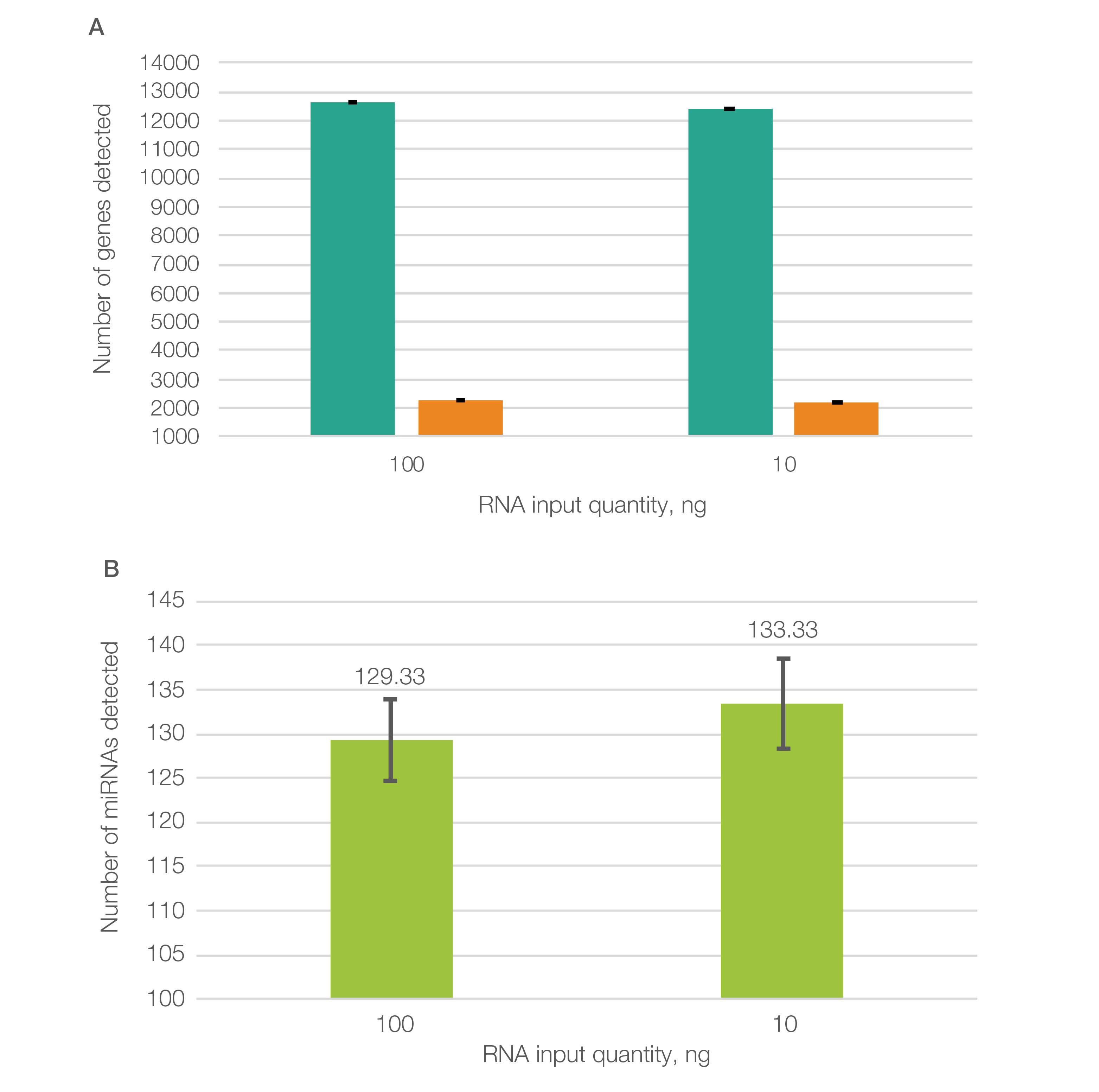
Fig. 1. Libraries prepared using the SEQuoia Complete Kit produced consistent sequencing results when run on the Element AVITI System. A, a similar number of protein-coding (■) and non–protein-coding (■) genes were detected with low (10 ng) and high (100 ng) quantities of input RNA. At least 5 reads were required to consider a transcript as detected. B, over 100 different miRNAs were detected (at ≥5 reads) on the Element AVITI System with both low (10 ng) and high (100 ng) quantities of input RNA. Note: Due to differing platform requirements, RNA-Seq libraries prepared using SEQuoia Kits to be run on the Element AVITI System were converted to compatible libraries using the Element Adept Library Compatibility Kit v1.1.
In evaluations of library performance using the SEQuoia Express Kit, results showed that data from the RNA-Seq libraries were highly correlated between the Singular G4 sequencing platform and a leading commercial benchtop sequencer (Figure 2; Kumar et al. 2023). Moreover, more long transcripts were detected in the data generated by the G4 Sequencing Platform overall, for both low (10 ng) and high (100 ng) input RNA.
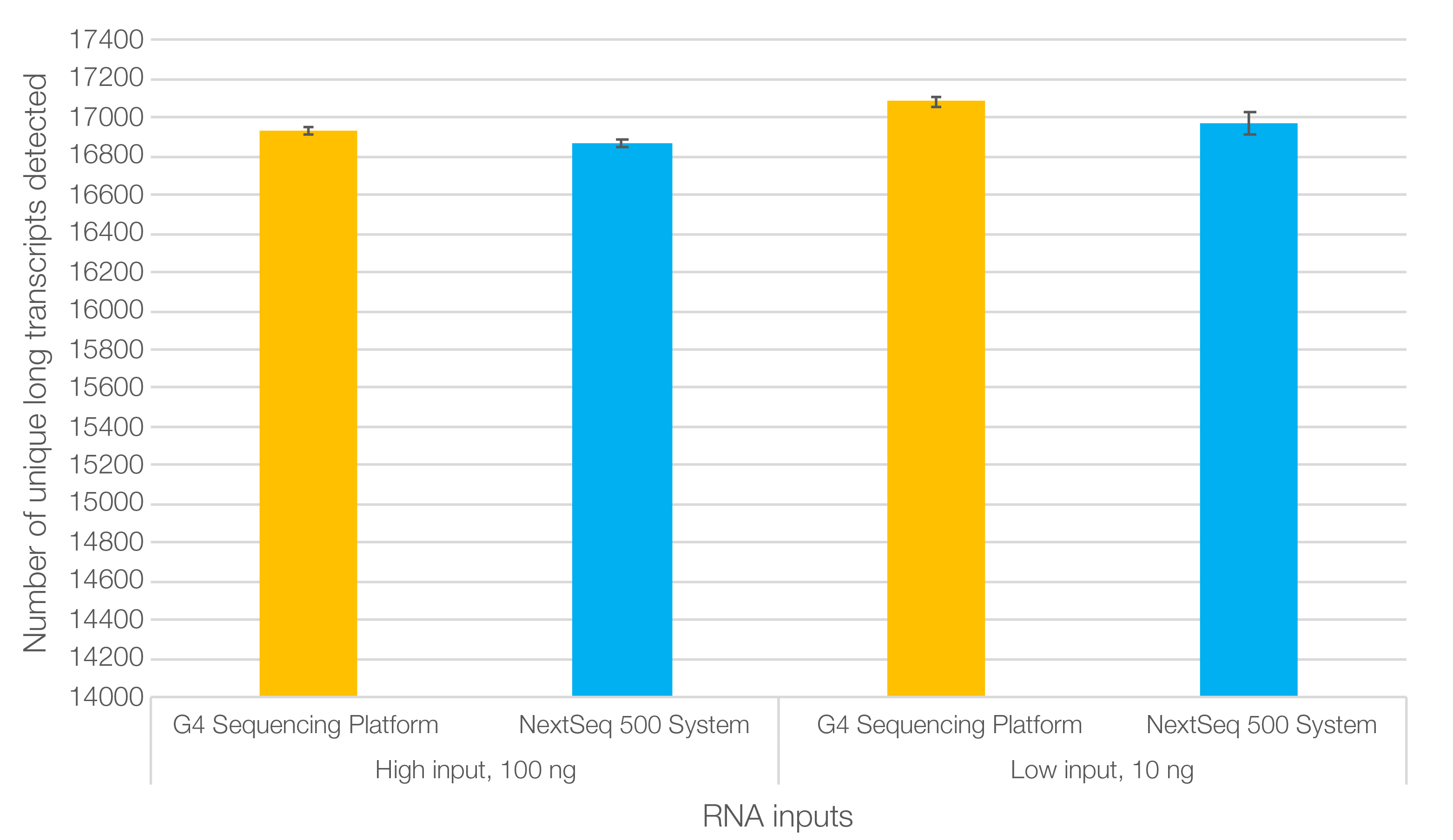
Fig. 2. SEQuoia Express RNA libraries run on the Singular G4 Sequencing Platform enable detection of more long RNA transcripts on average than the Illumina® NextSeq 500 System. Note: To enable the use of the SEQuoia Complete and SEQuoia Express Kits with the Singular Genomics G4 System, the RNA-Seq libraries were amplified with Singular Genomics Dual Index PCR Primers before being run on the sequencer.
Thus, SEQuoia Kits are readily suited to transcriptome profiling for a range of sample types with varying RNA input, and are compatible with new short-read sequencing platforms, ensuring the transfer of existing workflows to these new instruments or the development of new ones.
Not only can users bridge the gap to more affordable sequencing, but they can also obtain robust data that rivals the accuracy of a leading commercial benchtop sequencing platform. Depending on the SEQuoia Kit used, researchers can obtain the full transcriptome profile or access a more rapid library prep workflow of only 4 hours. This strategy can offer researchers more complete information from their samples at lower cost and effort and also enables access to high-quality data even if highly degraded RNA from FFPE samples is used.
Further Innovation is Needed to Address RNA-Seq Challenges
While new sequencing platforms and robust reagents are a boon to the adoption of applications that utilize RNA-Seq technology, many other aspects upstream and downstream of this process still require innovation. RNA-Seq is also part of a larger molecular analysis workflow. For example, pairing RNA-Seq with digital PCR and qPCR allows the confirmation of a broader discovery process using discrete smaller panels that are quick to implement in a large group of samples, once the relevant targets for a biological process are identified.
Some key areas where commercial focus could benefit or even transform RNA-Seq workflows are:
- Eliminate bioinformatics bottlenecks by providing integrated computational tools and resources aligned with new sequencing technologies for small and long RNAs
- Improved RNA extraction reagents for stable, high-quality RNA at sufficient yield would reduce the risk of degraded RNA from sample handling
- Sample pooling to run up to 96 libraries in a single tube to dramatically improve capacity
- Automated and streamlined RNA-Seq library preparation for higher throughput and standardization
- RNA-Seq data confirmation using orthogonal molecular technologies, such as digital PCR and qPCR
- Expansion of reference genomes for non-mammalian species, such as common crop plants and pathogens
- Overcome the affordability barrier to greater sequencing depth in the billions — dramatically increasing the number of reads per sample could identify low-level, previously undetectable transcripts
- Using single-cell RNA-Seq to determine cell-type-specific outcomes in the diverse heterogeneous tissue microenvironment aids developmental biology, neuroscience, and oncology
- The development of robust, cost-effective long-read sequencers capable of handling large fragments up to 1000 bp or more, to evaluate more of the RNA transcriptome, and fusion proteins, which short-read sequencers cannot detect (Gray et al. 2022)
RNA sequencing has revolutionized the era of RNA analysis in less than a decade, allowing research to forge into the unknown aspects of the transcriptome and delineate fundamental biological processes and disease states. Improving access to this unique technology in the next decade will help realize its full potential in both biological and translational research.
Visit our website to learn more about SEQuoia Kits and their use with the Singular Genomics G4 and Element AVITI Systems.
References
Chen H et al. (2022). Identification of differentially expressed genes at the single-cell level and prognosis prediction through bulk RNA sequencing data in breast cancer. Front Genet 13, 979829.
Gray ER et al. (2022). Ultra-sensitive molecular detection of gene fusions from RNA using ASPYRE. BMC Med Genomics 15, 215.
Kumar N et al. (2023). RNA sequencing on the Singular Genomics G4 Sequencing Platform using SEQuoia library preparation chemistries. Bio-Rad Bulletin 3464.
Perike S et al. (2022). Comprehensive capture of FFPE RNA for RNA sequencing using a continuous synthesis chemistry and post-library ribodepletion. Bio-Rad Bulletin 3421.
Mu T et al. (2020). Embryonic liver developmental trajectory revealed by single-cell RNA sequencing in the Foxa2eGFP mouse. Commun Biol 3, 642.
Stichel D et al. (2019). Routine RNA sequencing of formalin-fixed paraffin-embedded specimens in neuropathology diagnostics identifies diagnostically and therapeutically relevant gene fusions. Acta Neuropathol 138, 827-835.
Xia Y et al. (2022). Integrated DNA and RNA sequencing reveals drivers of endocrine resistance in estrogen receptor–positive breast cancer. Clin Cancer Res 28, 3618–3629.

