Multiplex Digital PCR Enables Tracking of Treatment Efficacy Using Liquid Biopsies
Disease-associated mutations, especially those linked to cancer, can be prognostic of outcome and inform treatment choices. Traditional tissue biopsies, however, limit our ability to implement such personalized treatments. These invasive surgical procedures can be used to genotype a tumor prior to treatment, but they are not amenable to serial tracking of treatment efficacy or disease progression. Furthermore, for some cancers, such as metastatic breast or prostate cancer, or for tumors that are not easily accessible, tissue biopsies can prove impossible.
By using cell-free DNA (cfDNA) to quantify cancer markers in blood or other bodily fluids, liquid biopsies can eliminate the need for invasive surgical procedures. However, the most commonly used approaches to quantify cfDNA, next-generation sequencing (NGS) and quantitative real-time PCR (RT-qPCR), have their own challenges.
Both NGS and RT-qPCR struggle to detect mutations present at <5% and are incompatible with often limited sample amounts. NGS requires complex workflows with long turnaround times and high costs while the relative nature of RT-qPCR makes it susceptible to PCR inhibitors often found in clinical samples.
Numerous studies have now shown that Droplet Digital™ PCR (ddPCR™) overcomes these challenges. By partitioning samples into droplets and using end-point PCR to amplify targets of interest, ddPCR enables absolute and highly sensitive nucleic acid quantification that is less susceptible to PCR inhibition than RT-qPCR and less costly and complicated than NGS workflows.
Prospective studies have used ddPCR to quantify driver and resistance mutations associated with tumors such as lung cancer (Sacher et al. 2016), breast cancer (Olsson et al. 2015), and melanoma (Chang et al. 2016) and have reported positive predictive values of up to 100%, turnaround times of 2–3 days, and a significant increase in sensitivity.
ddPCR assays for metastatic melanoma, for example, have been found to be 42% more sensitive than traditional LDH assays (Chang et al. 2016), and breast cancer recurrence could be identified up to three years earlier using ddPCR liquid biopsies than with traditional methods (Olsson et al. 2015). Following a highly successful clinical trial (Sacher et al. 2016) the Dana-Farber Cancer Institute and Brigham and Women’s Cancer Center now offer a Droplet Digital PCR–based liquid biopsy test to eligible non-small cell lung cancer patients.
The Dana-Farber study demonstrated that treatment response can be monitored by tracking patient-specific mutations in serial liquid biopsies. The hope is that in the near future, such information could be used to detect minimal residual disease (MRD) post-treatment, to identify recurrence and inform treatment changes, and to assess treatment resistance at the earliest time points, when disease is most treatable (Figure 1).
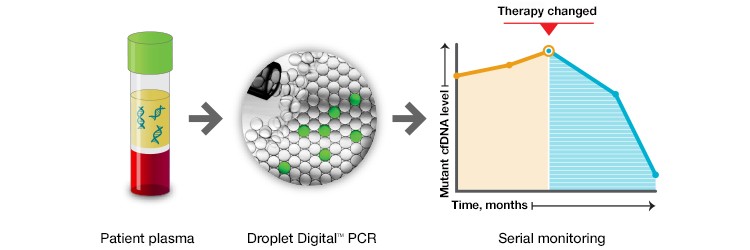
Fig. 1. Taking advantage of the high sensitivity of ddPCR and its simple and economical workflow, patient response to treatment can be monitored using serial liquid biopsies. Such approaches could be used in the near future to detect minimal residual disease, disease recurrence, or loss of treatment efficacy.
Here we demonstrate how ddPCR can be used to screen for several KRAS mutations associated with colorectal cancer (CRC) in a single reaction (Shelton et al. 2014).
Multiplex KRAS Mutation Assays Detect Mutations Present at 0.2%
The majority of CRC-associated KRAS mutations occur in codons 12 (~82%) and 13 (~17%) of exon 2. These mutations are predictive of a negative response to anti-epidermal growth factor receptor (EGFR) therapy and can thus inform treatment. Bio-Rad’s ddPCR KRAS G12/G13 Screening Kit allows screening for seven actionable codon 12 and 13 KRAS mutations as well as wild-type KRAS in a single reaction (Figure 2A).
FAM-labeled mutant probes and HEX-labeled wild-type probes result in four possible populations in a 2-D fluorescence plot (Figure 2B): (i) FAM- and HEX-negative events resulting from droplets that do not contain the KRAS gene; (ii) HEX-positive events, resulting from droplets that contain wild-type KRAS; (iii) FAM-positive droplets, that contain one or more of the assayed KRAS mutations; and (iv) FAM/HEX double-positive droplets that contain both wild-type and mutant KRAS.
The QX200™ Droplet Digital PCR System’s software quantifies these events and provides both an absolute number of copies and their fractional abundance. Mutants present at 0.2% abundance can be easily quantified (Figure 2C).
One key advantage of using ddPCR is that quantification is not relative. Because Droplet Digital PCR counts the absolute number of events, it is possible to determine the limit of detection and the minimum input required to attain a desired sensitivity (Figure 2D). This also allows for more accurate comparison of tumor burden during serial monitoring or when comparing treatment efficacy in different patients because measured fractional abundances are not relative.
KRAS Screen 7-Plex Assay Seven Mutant (FAM) and One Wild-Type (HEX) Probes
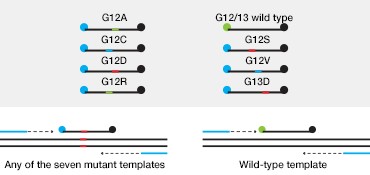
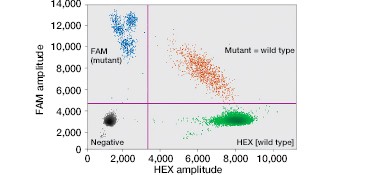
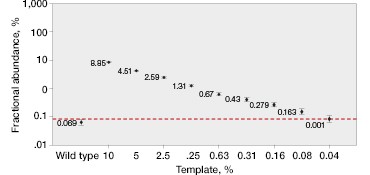
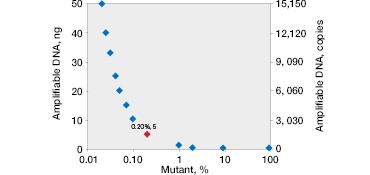
Fig. 2. Multiplexed single-well detection of seven actionable KRAS mutations. A, Bio-Rad’s ddPCR KRAS G12/G13 Screening Kit allows screening for seven common activating KRAS mutations and wild-type KRAS in a single reaction. B, multiplex assays result in four possible populations in a 2-D scatter plot. C, dilution series using G12D gDNA and two wells shows that KRAS mutations can be detected at abundances of 0.2%. D, absolute quantification allows rigorous quantification of detection limits.
Multiplex Detection of KRAS Mutations in cfDNA from Patient Samples
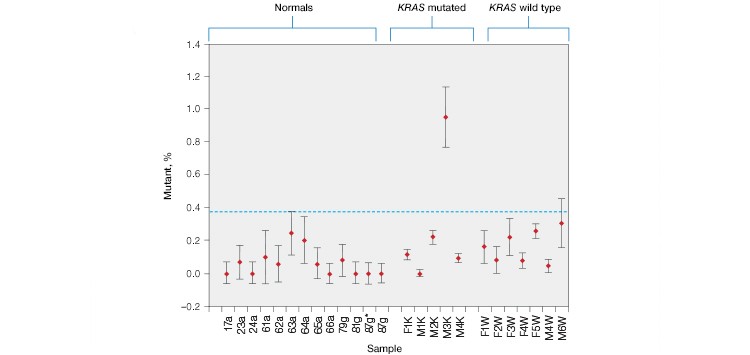
Using the ddPCR KRAS G12/G13 Screening Kit, KRAS mutations were detected in cfDNA patient samples (Figure 3A). Untested plasma samples from patients who had previously been classified as KRAS mutation positive or negative by FFPE genotyping were purchased from Conversant Bio and ProMedDx.
As expected, KRAS mutations were not detected in plasma from wild-type control patients or in samples from CRC patients that were FFPE genotyped as KRAS wild-type before treatment. KRAS mutations were detected in one out of the five KRAS mutation–positive samples, indicating that this patient’s treatment was likely unsuccessful. Using individual KRAS mutation assays, we were able to identify the M3K patient sample as KRAS G13D (Figure 3B).
A
B
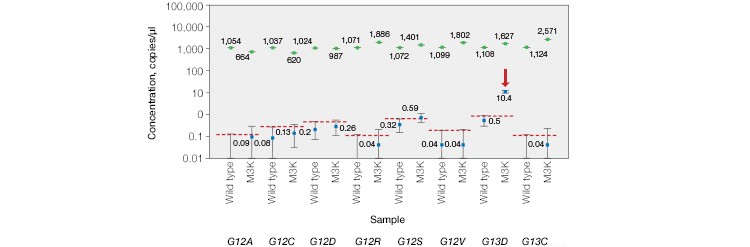
Fig. 3. Multiplex detection of KRAS mutations in patient cell-free DNA samples. A, the ddPCR KRAS G12/G13 Screening Kit detected KRAS mutations in 0/12 normal, 0/7 non-KRAS, and 1/5 KRAS-positive patient plasma samples. B, further analysis using individual KRAS mutation assays identified the M3K patient sample as KRAS G13D. Positive calls were made using nonoverlapping 95% confidence interval error bars. 87g*, sample repeat; green, wild-type KRAS; blue, mutant KRAS.
Conclusion
Using Droplet Digital PCR, actionable mutations present at frequencies as low as 0.2% can be quantified in cfDNA patient samples using a simple, fast, and economical workflow that is easily implemented in diagnostic laboratories. By multiplexing, several known cancer driver mutations can be assayed in a single reaction.
In addition to the ddPCR KRAS G12/G13 Screening Kit used in this study, Bio-Rad provides kits that enable screening for common NRAS and BRAF mutations and for KRAS Q61 mutations. When necessary, a tumor can be more specifically genotyped by using individual assays that can be designed in-house or purchased from Bio-Rad.
Droplet Digital PCR is equally impactful for cancers that have a large number of driver mutations. In these cases the more global view afforded by whole-genome NGS can be utilized to identify patient-specific mutations and targeted, more economical Droplet Digital PCR can be used to track these biomarkers throughout treatment.
References
Chang et al. (2016). Sensitivity of plasma BRAFmutant and NRASmutant cell-free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Molecular Oncology 10, 157–165.
Olsson et al. (2015). Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Molecular Medicine 7, 1,034–1,047.
Sacher et al. (2016). Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncology 2, 1,014–1,022.
Shelton et al. (2014). Droplet Digital PCR: Multiplex screening of KRAS mutations in cell-free DNA colorectal cancer samples. Bio-Rad bulletin 6679.
First posted on genengnews.com on October 1, 2016.

