Cancer remains one of the leading causes of death in the developed world. Tremendous advances in the diagnosis and treatment of malignant neoplasms have reduced the mortality rate. Statistics from the National Cancer Institute (NCI; cancer.gov/about-cancer/understanding/statistics) indicate that the overall cancer death rate fell by 13% from 2004 to 2013. Despite these advances in treatments and earlier detection, cancer still remains a major challenge in the healthcare field.
An Increasing Cancer Epidemic
The baby boomer generation is the result of the dramatic population increase from 1946 to1964, and the members of this group are currently in their mid-fifties or older. Lifesaving medical interventions have significantly increased the average human lifespan, meaning a greater proportion of the population is living longer than in any previous generation. This phenomenon, dubbed the silver tsunami, will challenge our healthcare infrastructure and is further compounding the urgency for effective cancer treatments (Bluethmann et al. 2016). At the same time, many aging populations increasingly suffer from chronic illness. So, while we are living longer, many of our later years are plagued by chronic illnesses. Most cancers are known to occur at any point in our lifespan, but many have an age-related onset. The NCI reports that just over a quarter of all cancer diagnoses occur between the age of 65 and 74, and an astounding 76% of all cancer diagnoses occur after the age of 55 (Figure 1). This sets the course for an oncoming wave of cancer patients who will require medical interventions to save their lives.
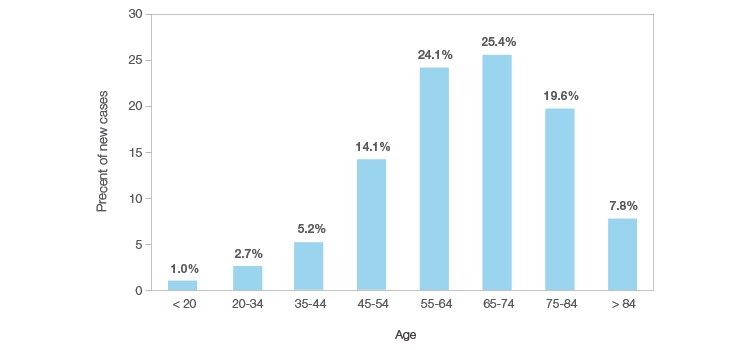
Fig. 1. Percent of new cancer diagnoses by age group. Bar plot showing the percentage of all new cancer cases by age group. Data are from a statistical analysis of the NCI’s Surveillance, Epidemiology, and End Results (SEER) program from 2007 through 2011 (seer.cancer.gov). Data borrowed from Age and Cancer Risk originally published by the National Cancer Institute.
This need is leading researchers to seek out new avenues for interventions in cancers and identify new biomarkers for the detection of neoplasms. One such area of active investigation is the field of long noncoding RNA (lncRNA) expression, which has the potential to serve both these needs. lncRNAs are a class of RNA species that do not encode proteins yet have functional roles in our biology. There are thousands of known long noncoding RNAs, and they outnumber protein coding genes in the human genome. Despite their prevalence, only a few hundred of these genes have well-defined biological functions. This is a treasure trove of genomic targets yet to be explored in the field of cancer biology.
A Peculiar RNA Discovery
RNAs that do not encode proteins are not a new concept to genetics. We have long known about other classifications of such RNAs, including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and small RNAs such as microRNAs. These RNAs are derived from genes with a defined structure and transcript products.
For many years it has perplexed researchers how humans can have nearly 3 billion bases in their genome and yet a mere 2% of them code for the entirety of their proteins. Early observations showed that many RNA transcripts are produced from a large proportion of the genome between protein coding genes. However, these were initially dismissed as spurious transcriptional events — simple background noise produced from leaky polymerases in the messaging to create proteins. While relegating these noncoding RNA as transcriptional static was a convenient explanation, and one that fit well into the widely accepted central dogma of molecular biology (DNA → RNA → Protein), some researchers could not accept that these transcriptional messages were without meaning or function. They sought to find order in this noise and examine if there might be some deeper complexity to our genome.
In 1988, a group of scientists from the Howard Hughes Medical Institute reported a “peculiar mRNA” known as H19. They noted that despite having all the characteristics of a protein coding gene (transcriptional start site, introns and exons, and polyadenylation) there was no discernible protein made by this gene (Brannan et al. 1990; Jarroux et al. 2017;Pachnis et al. 1988. This fascinating finding led to further scrutiny of the function of H19. The expression of this gene was found to be associated with ploidy (number of genome copies per cell). High levels of H19 are found in cells that are diploid (have two genome copies), while in cells that have greater than two genome copies, the levels of H19 are reduced. The conclusion was that expression of H19 correlates with the amount of genomic DNA present in the cell. Since cancer cells often display major genetic aberrations and chromosomal duplications, H19 has been associated with many different cancers in many studies. The discovery of a gene having all the appearances of a protein coding gene but not coding for one captured the imagination of the RNA biology field.
In the years following the discovery of H19, researchers in the long noncoding RNA field found several other examples of noncoding genes that have biological relevance. The technological advancement of massively parallel nucleic acid sequencing supporting RNA-Seq in subsequent years unleashed a mass of discovery for long noncoding RNAs. The role of lncRNAs in cancer became an active discussion in the cancer field and many cancer-associated lncRNAs were interrogated to better understand their function in the disease.
Long Noncoding RNAs as a Driver of Cancer
One of the major challenges in the lncRNA field has been ascribing function to these genes and understanding how they impact our biology. In addition to H19, several other genes were identified as being long noncoding RNAs and have been found to be differentially expressed in various cancers. Rather than relating to ploidy, these genes serve a wide array of other cellular functions related to cancers. These RNAs function in a variety of ways from activating or repressing transcription to acting as a scaffold for recruitment of epigenetic modulating complexes. As a result, a new class of RNA, designated long noncoding RNA, was established and includes RNA longer than 200 nucleotides. Though this was quite a broad definition, it still allowed for a framework to research this new class of RNA.
Table 1, with just a few examples of cancer-related lncRNA, highlights the diverse functions these RNAs can have. They can hybridize through nucleic acid base pairing, but far more importantly, they can utilize their secondary and tertiary structures to their advantage to interact with other molecules. This versatility of interactions within the cell and the sheer volume of these genes within our genome begs the question of how they eluded detection for so long.
Table 1. Diverse functions of lncRNA in cancer.
| Type of lncRNA | Example | Cancer Phenotype |
| Antisense lncRNA | ANRIL | Antisense to transcripts in the CDKN2B locus, which has many disease-associated genes |
| miRNA decoy | HULC, PTENP1 | Modulate miRNA function by physically binding and sequestering them |
| Splicing modulator | ABALON | Cause alternative splicing of other RNAs |
| Enhancer of transcription | H19, KCNQ1OT1 | Drive the expression of oncogenes |
| Epigenetic modulator | HOTAIR, ANRIL | Recruit chromatin modifying factors |
The previously mentioned technological advances that have enabled the interrogation of the entire transcriptome highlight some challenges to the research of long noncoding RNAs. For one, these transcripts are present at a magnitude of order less than protein coding genes on average. Their expression is also typically seen in the context of cell-specific or temporal cell differentiation. Locating and studying such targets requires sensitive detection methods that can also operate on potentially limited sample material. Despite the challenges of measuring the presence of lncRNAs, their characteristics indicate that (on average) they are very potent and operate in very particular biological states. These characteristics make them very attractive druggable targets and as points of intervention for modulating a disease.
The Potential of lncRNA in Cancer Diagnosis and Intervention
Cancer screening methodologies that include noninvasive biological matrices such as blood, urine, or saliva have long been sought out as clinical diagnostics. These tests have utility for both detection of a disease and routine monitoring treatment efficacy. The challenge is often finding an analyte that is specific and sensitive to a particular cancer phenotype (Bolha et al. 2017). lncRNAs are often expressed in very tissue specific contexts, and those that are differentially expressed in certain cancer types could serve as new molecular biomarkers for cancer (Figure 2). Many lncRNAs are also found to be relatively stable circulating in body fluids; this potentially confers the ability to detect lncRNA biomarkers through noninvasive means (Dai et al. 2017; Xie et al. 2018). Testing of lncRNAs is also potentially more informative than testing of DNA since the latter is informative for disease risk while RNA tests can offer information about the current state of a patient through quantification of expression levels. The FDA has already approved the use of a diagnostic test for the lncRNA PCA3 as a marker for prostate cancer in urine (Rönnau et al. 2014; Smolle et al. 2017), and other similar noninvasive diagnostics are being explored for other cancer types.
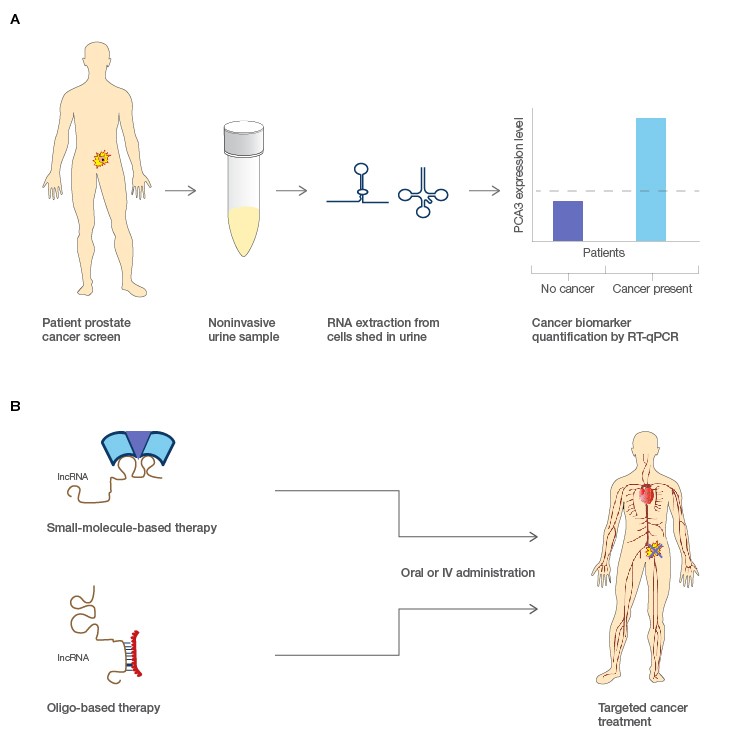
Fig. 2. Potential applications for lncRNA in molecular diagnostics and pharmacological treatment of cancers. A, lncRNA biomarkers associated with prostate cancer could be extracted from urine samples and evaluated for their expression levels as a simple noninvasive cancer diagnostic. B, small molecule or antisense oligo treatments could target the function of lncRNA and modulate downstream protein coding genes that cause disease. Figure modified from Prensner and Chinnaiyan 2011.
Long noncoding RNA are also attractive targets from a pharmaceutical perspective because they can directly modulate disease-related gene expression. They could potentially be targeted through the use of small molecule compounds that interact with the tertiary structure of the lncRNA or through the use of antisense oligos that base pair with the lncRNA sequence itself. This latter concept is one that has renewed great interest in translational research in the lncRNA field. The explosion of lncRNA research has reinvigorated RNA biology as an attractive target for pharmacology.
For treatment of cancers, there is interest in developing treatments targeting long noncoding RNAs to modulate their function. These could take the form of many potential interventions such as screening small molecules that interact with the structure of the RNA and modulate their function directly or regulate their expression indirectly. Recently, an antisense oligo known as nusinersen (commercially known as Spinraza) was approved by the FDA for the treatment of a rare but debilitating and often deadly disease known as spinal muscular atrophy (SMA). This drug has demonstrated that we can use a nucleic acid to directly modulate the splicing of an RNA molecule to cure or dramatically slow the progression of a disease. The success of this antisense oligo drug has demonstrated the possibility to treat other diseases such as cancers by taking a similar approach — targeting long noncoding RNAs that contribute to tumor progression. Because there are tens of thousands of long noncoding RNAs in our genome, there is a rich pool of potential candidates to explore in cancer regulation and target for treatments for either oligo based treatments or conventional small molecule compounds.
The coming years will be an exciting era in genetic research as we discover new biological mechanisms and functions for noncoding RNAs. Our improved understanding of this class of genes should provide insight into novel treatments for cancers.
Bio-Rad offers a complete solution for simple and sensitive expression analysis of long noncoding RNAs. See how we can help you develop and simplify your lncRNA workflow.
References
Bluethmann SM et al. (2016). Anticipating the “silver tsunami”: Prevalence trajectories and comorbidity burden among older cancer survivors in the United States. Cancer Epidemiol Biomarkers Prev 25, 1,029–1,036.
Bolha L et al. (2017). Long noncoding RNAs as biomarkers in cancer. Dis Markers 2017, 7243968
Brannan CI et al. (1990). The product of the H19 gene may function as an RNA. Mol Cell Bio 10, 28–36.
Dai Q et al. (2017). Circulating long non-coding RNAs as biomarkers of acute myocardial infarction. Non-coding RNA Investig 1, 4.
Jarroux J et al. (2017). History, discovery, and classification of lncRNAs. Adv Exp Med Biol 1008, 1–46.
Pachnis V et al. (1988). The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J 7, 673–681.
Prensner JF and Chinnaiyan AM (2011). The emergence of lncRNAs in cancer biology. Cancer Discov 5, 391–407.
Rönnau CG et al. (2014). Noncoding RNAs as novel biomarkers in prostate cancer. Biomed Res Int 2014, 591703.
Smolle MA et al. (2017). Current insights into long non-coding RNAs (lncRNAs) in prostate cancer. Int J Mol Sci 18, E473.
Xie Y et al. (2018). Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non‐small cell lung cancer. Mol Oncol [published online ahead of print March 5, 2018]. Accessed March 31, 2018.
Spinraza is a trademark of Biogen MA Inc.

