Unsure whether to choose quantitative PCR (qPCR) or Droplet Digital™ PCR (ddPCR) for gene expression analysis? Bio-Rad’s new objective study comparing the two methods shows that the best choice depends on your targets and experimental design. Although ddPCR produced higher quality data overall, especially for low-abundance targets, qPCR can offer a throughput advantage when working with more abundant targets.
Introduction
Gene expression analysis is foundational to molecular biology, helping researchers decode how genes respond to different biological conditions, treatments, or disease. For years, quantitative PCR (qPCR) has been the method of choice, valued for its speed, sensitivity, ease of use, and well-established guidelines for publication-ready results. Droplet Digital PCR (ddPCR) has emerged as a powerful complementary platform, offering high resolution and exceptional precision, especially for low-abundance targets and subtle-fold changes. However, broader adoption of ddPCR technology has been limited by the lack of standardized frameworks that show how results can be comparable between studies.
Here, we share key findings from a new gene expression study that puts both technologies to the test, with a side-by-side comparison of singleplex and multiplex assay formats of the qPCR and ddPCR systems. The results reveal that each platform offers unique strengths depending on experimental design, sample type, number of targets, and the level of expression change being measured. While qPCR is an established technique for gene expression of abundant targets and normal RNA input, the ddPCR platform can deliver higher sensitivity, better precision, and easier multiplexing that is particularly valuable for low-expressing or limited samples. By selecting the right tool for the task, researchers can unlock more robust, accurate, and comprehensive gene expression analysis — paving the way for improved reproducibility and deeper biological insight.
Comparative Data from a Gene Expression Study
Enhancing Gene Expression Analysis: Leveraging ddPCR Technology Alongside qPCR
In this study, the authors evaluated the CFX Opus Real-Time PCR System and the QX600™ddPCR System to explore which platform maximized precision, sensitivity, and robustness in gene expression workflows depending on experimental needs.
Experimental Design and Dual-Platform Setup
To ensure data comparability, identical cDNA samples and primer sets were used for both qPCR (CFX Opus System) and ddPCR (QX600 System) analyses. Multiplex panels included BCL2 and GADD45A as target genes (with treatment of cisplatin). ACTB and PGK1 were selected as reference genes using the PrimePCR Reference Gene Panel to serve as normalization controls across both platforms. qPCR data was analyzed via CFX Maestro Software using the ΔΔCq method, while the ddPCR assay employed QX Manager Software for high-resolution analysis. The use of consistent assay designs in the cisplatin model enabled a side-by-side performance evaluation. For more information on experimental setup see Bio-Rad Bulletin 3867 (coming soon).
Application Study Insights
qPCR offers a broad dynamic range, which makes it highly suitable for samples with widely varying target concentrations. Its rapid cycling protocols support high-throughput workflows with fast turnaround times, making it ideal for experiments requiring timely results. In addition, qPCR benefits from mature analysis pipelines and well-established publication standards, enabling consistency and comparability across studies.
While qPCR remains a go-to method for many gene expression studies, this study explored how ddPCR assays can provide distinct advantages in specific experimental contexts.
Improved Sensitivity for Low-Abundance Targets
ddPCR technology quantifies gene expression without relying on Cq values, standard curves, or amplification efficiency. This makes it particularly advantageous when studying low-abundance targets, subtle fold changes, or when reference gene stability is variable, such as in stem cell growth and differentiation studies.
As shown in Figure 1, both qPCR and ddPCR assays effectively measured gene expression, including low-abundance target BCL2. While qPCR successfully detected BCL2, it did not identify a statistically significant change in fold expression. In contrast, ddPCR technology detected BCL2 and resolved a significant fold difference with tighter error bars, demonstrating greater precision and sensitivity for subtle expression changes.
Singleplex qPCR and ddPCR Assays
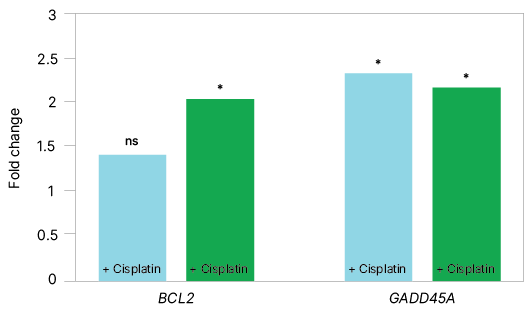
| Fold Change | ||
| Target Gene | qPCR | ddPCR |
| BCL2 | ns | 2.07 |
| GADD45A | 2.36 | 2.3 |
Multiplex qPCR and ddPCR Assays
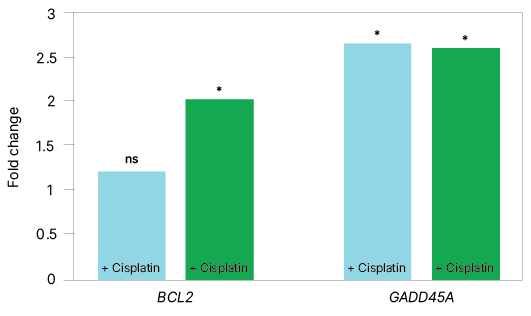
| Fold Change | ||
| Target Gene | qPCR | ddPCR |
| BCL2 | ns | 2.03 |
| GADD45A | 2.66 | 2.6 |
Fig. 1. Comparison of target gene expression data using singleplex and multiplex assays on qPCR and ddPCR platforms demonstrates that ddPCR assays can detect twofold gene expression differences. Bar graphs represent target gene expression data acquired using singleplex and multiplex assays across both qPCR (■) and ddPCR (■) platforms, with and without cisplatin treatment at 24 hours. qPCR data analysis was performed using a CFX Maestro Gene Study with data obtained from the CFX Opus Real-Time PCR System, whereas ddPCR data analysis was performed using a QX Manager Gene Study with data obtained from the QX600 Droplet Digital PCR System. Significant gene expression fold changes (greater than ±twofold) were detected on both platforms. ns, not significant.
Multiplexing Efficiency and Higher Orders of Multiplexing
Both qPCR and ddPCR platforms demonstrated robust performance in singleplex and multiplex formats, as shown in Figure 1. A key advantage of Droplet Digital PCR is its simplified multiplex development. Because ddPCR technology is not dependent on amplification efficiency, it supports accurate multiplexing with less input material and minimal optimization, maintaining accuracy in more complex assays, even when multiple targets are added.
In contrast, conventional qPCR multiplexing typically requires assay validation to ensure matched performance across several targets. However, this requirement is mitigated when using Bio-Rad PrimePCR Assays, which are pre-optimized for annealing temperature and amplicon length. This feature is especially valuable when working with limited or precious samples. Additionally, by using PrimePCR Assays, designed to work off the shelf across both qPCR and ddPCR platforms, researchers can transition between technologies without re-optimizing assays, providing flexibility and streamlining workflow integration.
Enhanced Reproducibility and Precision
While both qPCR and ddPCR platforms generated similar expression patterns overall, the tighter error bars across replicates in ddPCR assays give it a clear edge in terms of statistical confidence, which is particularly beneficial for studies requiring high resolution or when measuring subtle changes in gene expression. Additionally, end-point measurements in ddPCR assays make it less susceptible to reaction inhibitors that can compromise qPCR efficiency and data precision.
Comparison of qPCR and ddPCR Technologies in Gene Expression Analysis
While qPCR remains highly effective for routine gene expression studies with moderate-to-high abundance targets, ddPCR technology expands detection capabilities, especially for low-level transcripts or challenging sample types. Together, these platforms offer complementary strengths: qPCR is optimal when target expression is robust and throughput is a priority, while the ddPCR platform provides added value in scenarios demanding precision, low sample input, or multi-target analysis in a single assay. This study highlights key considerations for selecting the appropriate method based on assay complexity, sample type, and expression level, laying the groundwork for more detailed guidance in future publications.
| Category | qPCR | Droplet Digital PCR |
| Assay compatibility | Easy transition of validated assays across platforms, thus reducing optimization efforts (e.g., with PrimePCR assays) | |
| Quantification method | Relative: ΔΔCq method; standard curves required during assay optimization | Absolute (copies per uL): no Cq or standard curve |
| Detection sensitivity | Best for moderate-to-high abundance targets (>100 copies): Targets with a Cq above 30 can show reduced reproducibility. Reliability begins to decline with a Cq above 35. | Detects low-abundance targets (down to 0.5 copies/μL) |
| Precision | Good precision for mid/high expression levels (>twofold changes) | Higher precision; smaller error bars and reliable detection of less than twofold differences |
| Reference genes | Essential for normalization | Dependent on experimental design—recommended for samples with varying input |
| Multiplexing | Needs validation and/or optimization to ensure all assays are amplified with near perfect efficiency | Ease of multiplexing without having to optimize for amplification efficiency |
| Sample input | More input required for low-expressing genes | Low sample input; ideal for limited material |
| Contaminant impact | Susceptible: May require extra assay optimization and specialized supermixes | Resilient due to end-point analysis |
| Best use case | Moderate to high-expression targets, known references | Low-expression targets, subtle changes, multiplex workflows |
Bio-Rad Continues to Innovate PCR Technology
Since pioneering qPCR in 1999, we have continually driven innovation by advancing both qPCR and ddPCR platforms. This commitment has positioned Bio-Rad as a leader in enabling precise and accessible gene expression analysis. The latest ddPCR systems reflect this legacy by delivering a streamlined workflow familiar to qPCR users—starting from standard PCR plates and removing labor-intensive steps previously associated with ddPCR assays.
The QX ContinuumTM Droplet Digital PCR System is a product of this ongoing innovation. This new platform delivers the hallmark performance of ddPCR technology with the familiarity of qPCR and offers a cost-effective solution with our simplest ddPCR workflow. With the ability to quantify up to four targets from a single sample well, its multiplexing capability is on par with that of the CFX Opus Real-Time PCR System. Additionally, its ddPCR technology offers the sensitivity needed to detect small changes in gene expression levels.
The QX Continuum Droplet Digital PCR System combines droplet generation, thermal cycling, and droplet reading in a single instrument.
Researchers can now achieve higher resolution and absolute quantification with minimal disruption to their existing protocols. This seamless integration ensures that both qPCR and ddPCR technologies evolve hand-in-hand, empowering scientists to harness the full power of digital PCR while maintaining the simplicity and efficiency they rely on.
Learn more about gene expression analysis.




