Watch this video to see how two scientists from the Southern California Coastal Water Research Project (SCCWRP), Yiping Cao and John Griffith, are working on a more efficient and accurate water testing method using Droplet Digital PCR technology.
A great day at the beach?
It’s a beautiful day at Corona Del Mar, a classic Southern California beach. Waves, blue and crystalline under a cloudless sky, roll in over a white sand beach. A child chases seagulls through the foam as her grandparents walk behind her. Today the waters are just as clean as they appear. No bright red signs or yellow ribbons block off the sand, warning beachgoers away.
It’s not always so. An hour and a half to the north, at another famous spot on the Southern California coast, the area near Malibu Pier was recently named one of 17 national “repeat offender” beaches by the Natural Resources Defense Council. This site earned that unfortunate honor for failing to meet water quality benchmarks more than 25% of the time. (The same report classed Newport Beach, near Corona Del Mar, as one of the country’s “superstar” beaches for its clean record.) Nationwide, about 10% of coastal water samples show bacteria counts exceeding levels the Environmental Protection Agency considers safe for swimmers. This amounts to 20,000 days per year when the red signs go up, signaling either a warning or full closure at one of the country’s beaches.
Even with these precautionary measures, the EPA estimates that around 3.5 million Americans get sick each year from polluted coastal waters. While rare illnesses are the ones that make the news, the usual culprits are—yuck alert!—the common viruses and bacteria found in human sewage that contaminates beach areas. These are the same bugs, the enteroviruses, adenoviruses, and noroviruses, that often cause food poisoning or the common cold. Most Americans don’t make the connection that the cause for many vacations cut short or ruined by sickness is a bad day at the beach.

Corona Del Mar is one of southern California’s most beautiful beaches. However, even pristine beaches like this one can become contaminated with microorganisms, especially during California’s rainy seasons, when sudden storms can wash pollution from the land into the water.
A small agency with a funny name
The sources of coastal water pollution depend largely on the nearby landscape and the human activities that take place there. Those can include leaky sewer lines, septic system and sewerage treatment overflows, runoff, pollution entering streams and rivers, and boats. “Here in Southern California everything is paved over,” says Dr. John Griffith, principal scientist and head of the Microbiology Department at the Southern California Coastal Water Research Project (SCCWRP). “Everything is engineered to prevent flooding, so when it rains, when there’s runoff, or when a fire hydrant breaks, it all runs down to the beach just as fast as it can.”
Griffith’s agency — everyone there pronounces its acronym as a word, “skwerp” — is tasked with a dual mission: study the causes and effects of what’s running down to the beach, and recommend ways to mitigate harm. As such, SCCWRP functions as an independent research institute providing and interpreting unbiased scientific information for policy makers and government agencies ranging from the EPA down to the local sewer district. Its location, close to one of Orange County’s wastewater treatment plants, the dry, paved-over bed of the Santa Ana River, and pristine Corona Del Mar beach, suggests its role in mediating between these manmade and natural water systems.
One of the main ways SCCWRP plays this role is by serving as a lab for improving water monitoring methods. “We’re interested in making sure we’re using the most advanced methods to protect public health from pathogens, primarily from human waste,” Griffith says. “That means we’re a major conduit for getting new technologies to the beach water quality monitoring community.”
A lagging and unspecific measure of fecal pollution
Across the United States, various groups monitor about 3,500 different coastal sites. Because environmental samples are complex and disease-causing pathogens such as viruses can be hard to detect, monitoring groups instead look for a more plentiful indicator organism. When monitoring water for fecal contamination, Enterococcus bacteria serve this purpose. Though generally harmless to humans on their own, the presence of Enterococci can indicate fecal pollution and the various pathogens that accompany it. Testing water samples to see if they grow Enterococcus cultures at a rate greater than 104 colony forming units (cfu) per 100 ml has proved a durable and simple method for detecting dangerous pollution.
However, Griffith and his colleague at SCCWRP, senior microbiologist Dr. Yiping Cao, see drawbacks to using cell culture as a method for monitoring coastal water quality. “The main issue is that it takes a whole day to get an answer,” says Cao. “The growing process takes 24 hours, so by the time you have an answer it’s the next day. The water stays open for the public to use, and the pollution event may have already passed by the time you know.”
Besides the delay in getting results, Cao also notes that, for all its benefits as an indicator, Enterococcus is not specific to the human waste that’s a particular public health concern. Because of these limitations, Cao has been investigating new approaches to water monitoring. “We’re looking for something that’s faster and will also measure organisms that are more specific to human fecal material,” she says.

Meredith Raith, a senior research technician at SCCWRP, prepares to take a sample from the Santa Ana River. The concrete culvert protects surrounding Orange County from major floods, but also channels waste from the land directly into coastal waters.
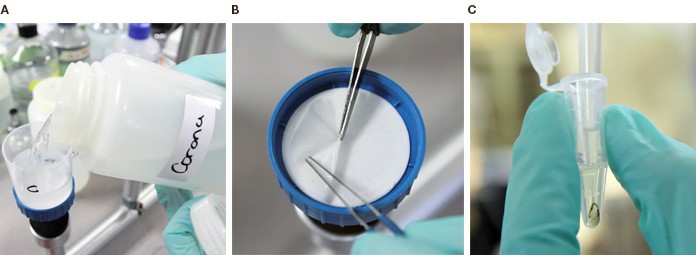
From the environment to the test tube: A. Water samples are put in a vacuum filter; B. The filters are then stripped of microorganisms, which are then lysed; C. The resulting genetic material is ready for PCR analysis.
qPCR came to the rescue, but…
Water quality researchers believe that checking for genetic markers offers a better measure of relevant contamination than growing bacterial cultures. As a method, quantitative real-time PCR (qPCR) can help solve both the speed and specificity issues associated with growing Enterococcus colonies in a dish. Using a qPCR assay gives quantitative results in a few hours, rather than a full day; also, researchers have designed qPCR assays to measure human-associated Bacteroidales, an additional indicator that increases the odds of detecting human contamination. In a shift from the generally used culture method, the EPA recently approved a qPCR protocol for Enterococcus for use in recreational water quality monitoring.
While Cao sees qPCR as an improvement, she nonetheless finds that this method poses its own limitations. “qPCR has its own shortcomings,” she says. “For one, it relies on an external standard [Enterococcus genomic DNA] for quantification. Depending on where you get your standard, it can cause high variability in your results.” A study Cao and her colleagues conducted found that among standards offered by various sources, this variability could produce up to half a log difference in results. This variability in standards becomes especially important once you consider the 3,500 different monitoring sites across the country, with measurements done by groups that might not all use the same standard. Such a range of results would be very hard to compare nationally, making it hard to decide when to close a given beach.
Inhibition is another issue with qPCR. This happens because environmental samples are so complex. That is, they can contain many different compounds depending on where and when they’re collected. “Some of these compounds can interfere with the PCR process, causing false negatives in qPCR,” says Cao. “Those are a big concern for public health protection. You always prefer to overprotect rather than under protect.”

Meredith Raith closes the Droplet Generator of SCCWRP’s Droplet Digital™ PCR (ddPCR) system. Yiping Cao, senior microbiologist at SCCWRP, cites the ability of ddPCR to offer absolute quantification of target genes, to allow multiplex analysis of different pathogens, and to minimize the reaction inhibition that can result from chemicals in complex environmental samples.
Water monitoring goes digital
In seeking to develop a better method for using genetic markers to monitor coastal water contamination, Cao and her colleagues have been investigating Bio-Rad’s Droplet Digital PCR (ddPCR™) technology. This PCR method utilizes the principle of sample partitioning to achieve a digital result through the statistical analysis of positive and negative partitions.
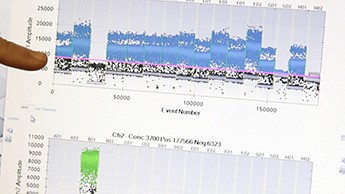
Taking an early look at a duplex assay. Cao’s assay measures both Enterococcus bacteria (blue channel) and a human fecal indicator organism (green channel).
Cao initially became interested in ddPCR because, unlike qPCR, it doesn’t rely on a standard curve to produce quantitative results. “We want to remove the variability caused by having an external standard,” she says. “Digital PCR provides absolute quantification so you get more reproducible data.” Absolute quantification of genetic material is possible by dividing the sample across approximately 20,000 uniform droplets, with each functioning as its own PCR chamber. The droplets are then read as either positive or negative; the fraction of positive droplets is translated by the software in a highly precise measurement of the template concentration.
Because digital PCR measures the end result of a reaction, rather than its kinetics, using ddPCR also helped with the issue of PCR inhibition. “The inhibition problem essentially went away,” says Griffith. “As long as there’s amplification it doesn’t matter if it’s in cycle 15 or cycle 35; we can detect it and use that result, so it’s much more accurate than it would otherwise be.”
As Cao learned more about ddPCR technology, she realized that she could measure both Enterococcus and human fecal-associated Bacteroidales at the same time, a result that had eluded her using qPCR multiplex methods because of competition for reagents. “With Droplet Digital PCR, you’re doing your reaction in very small volumes,” she explains. “Competition for reagents is minimized within the droplets, so that allows multiplexing.” That gives Cao the ability to detect the two key indicator organisms in the same reaction, saving both lab hours and reagents.
Monitoring that’s faster, cheaper, and on wheels

Using genetic analysis methods let water monitoring groups assess hazards in hours rather than the full day it takes using culture-based methods. ddPCR improves this further by allowing more accurate tests for multiple genetic targets at once, as well as the potential to analyze new genetic markers and better track pollution sources.
In the near term, Cao’s initial results have been promising, showing greater reproducibility and reduced reaction inhibition compared to qPCR. By measuring two indicators at once rather than in two separate runs, her ddPCR assay may also offer a significant per-sample cost saving over qPCR. This is no small consideration in an era when environmental agencies at all levels must deal with shrinking budgets while still carrying out their mandate to protect public health.
Cao has begun presenting her duplex ddPCR assay to her colleagues and assembling her data for publication. With further validation, she hopes that the assay may offer a useful alternative method for application in water quality assessment, particularly for fecal source identification. “We intend to train our member agencies who are interested in this new technology,” she says. “Our hope is that this assay becomes a valuable tool for monitoring agencies or researchers looking for more accurate and reproducible measurements.”
However, the duplex assay is not the only application ddPCR offers. “It’s a technology for the future. It really opens the door to a lot of research questions that we haven’t been able to answer before,” Cao says. Those include looking at other genetic markers that might better pinpoint the fecal sources of harmful pathogens or else target the pathogens directly. Getting a more accurate measure of fecal source–specific markers also might enable improved source tracking, letting water monitors prioritize pollution remediation.
For now, though, the immediate impact is the potential for better, faster information for swimmers. “If we could get the answer the same day, early in the day, that’s a really important thing,” says John Griffith. “We’d actually like to package this [ddPCR] technology into a mobile unit that telemeters data right back to the lab. Having that and the ability to measure both indicators at the same time would provide a much more definitive and actionable piece of information for public health protection.”
Disclaimer: The views expressed in this article are not necessarily those of the Southern California Coastal Water Research Project Authority or its member agencies nor an endorsement of a particular brand or technology.



