While the concepts behind digital PCR have been understood for about two decades, practical implementations of the technique have become available only in the past few years. Bio-Rad’s QX200™ Droplet Digital™ PCR system relies on sample partitioning by microfluidics to create 20,000 individual reaction chambers in each run, which results in an absolute measurement of target DNA without the need for a standard curve. Partitioning on this scale also minimizes competition between assays, allows for better multiplexing, and improves sample throughput.
Having the ability to obtain a truly “digital” measurement of target DNA opens the door for numerous applications. That the QX200™ ddPCR™ system accommodates both EvaGreen and probe-based detection chemistries makes even more applications possible. Though new and innovative uses for Droplet Digital PCR are being pioneered every day, here are some of the current applications for this exciting new digital PCR platform.
For more details, visit the Digital PCR Applications & Technologies page or download the complete guide to ddPCR Applications.
EvaGreen is a trademark of Biotium, Inc. Bio-Rad Laboratories, Inc. is licensed by Biotium, Inc. to sell reagents containing EvaGreen dye for use in real-time PCR, for research purposes only.
Quantifying Cancer Biomarkers
Realizing the full potential of cancer biomarkers to characterize disease states depends on detecting small but significant fluctuations in these key genetic variants. Using Droplet Digital PCR technology, researchers are now able to observe much finer quantitative distinctions among mutations, better pinpointing their potential roles in cancer.
Cancer-associated mutations often evade detection due to their low concentrations relative to the background of wild-type DNA in a given sample. With its high sensitivity, the QX200™ Droplet Digital™ PCR system can easily scale to quantify target concentrations as low as one out of 1,000,000 (0.0001%) total copies. What was previously undetectable with other methods can now be quantified with Droplet Digital PCR.
Detection of BRAF V600E Mutation
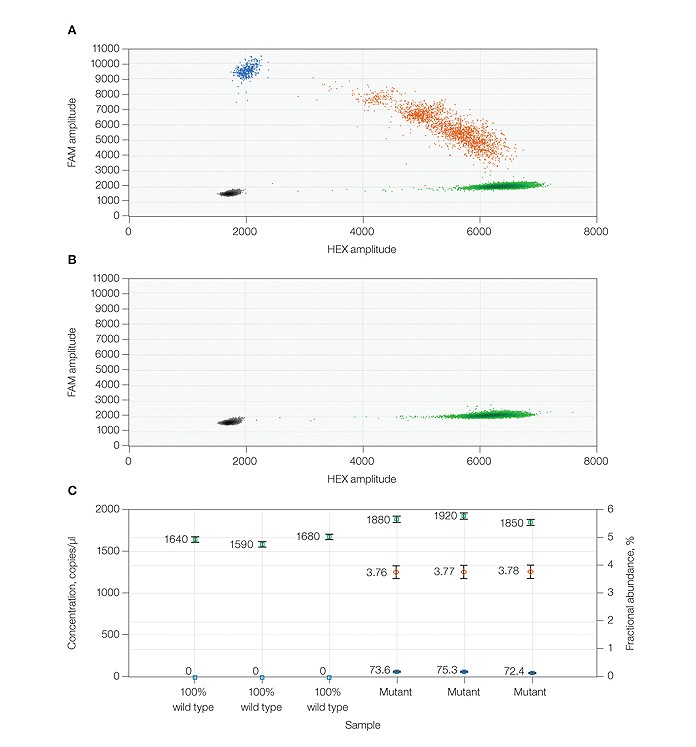
High sensitivity of the QX200 system allows quantification of BRAF V600E mutation in the presence of wild-type DNA using PrimePCR™ ddPCR™ assays. A, 2-D fluorescence amplitude plot generated by QuantaSoft™ software shows triplicate wells of a mixed mutant:wild-type-sample. The black cluster on the plot represents the negative droplets, the green cluster represents the droplets that are positive for wild-type DNA only, the blue cluster represents the droplets that are postive for mutant DNA only, and the orange cluster represents the droplets that are positivive for both mutant and wild-type DNA. B, 2-D fluorescence amplitude plot shows three replicates of a wild-type-only sample. C, fractional abundance plot shows the percentage frequency (orange markers) of the mutant DNA in a wild-type DNA background. The blue markers indicate the concentration of mutant DNA a wild-type DNA background. The blue markers indicate the concentration of mutant DNA (copies/μl) and the green markers indicate the concentration of wild-type DNA (copies/μl) in each of three replicate samples. All error bars generated by QuantaSoft™ software represent the 95% confidence interval.
Insight into Viral Reservoirs
Precisely quantifying viral load is crucial for characterizing disease states and developing and validating therapies. Viral reservoirs fluctuate, sometimes dropping to very low levels that are nonetheless significant. Obtaining exact and reproducible measurements of viral DNA or RNA often represents the difference between success and failure in studying many different pathogens.
The QX200™ Droplet Digital™ PCR system has the sensitivity to register extremely minute quantities of viral genetic material and distinguish it from complex mixtures. Investigators can obtain actual copy numbers of viral target RNA by generating complementary DNA through reverse transcription. Along with precision and reliability, the QX200 system also offers users the throughput needed to process samples with great efficiency.
Detection of Herpes Simplex Virus and β2 Microglobulin
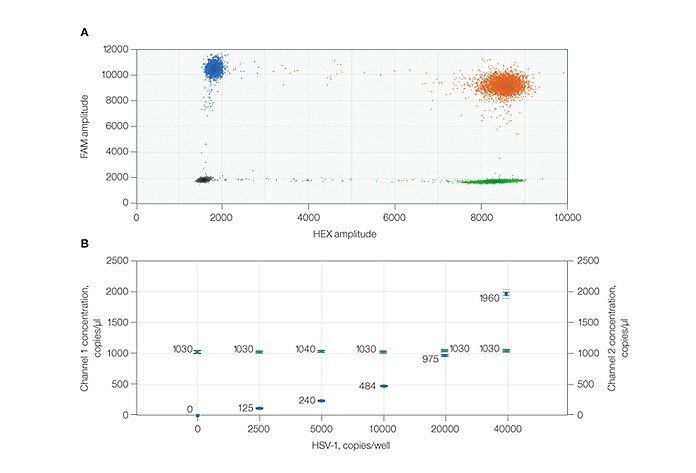
ddPCR™ technology enables precise and reproducible detection of herpes simplex virus 1 (HSV-1) and β2 microglobulin (B2M) targets in a duplex assay. A, 2-D fluorescence amplitude plot shows three merged replicate samples of 10,000 copies/well of HSV-1 duplexed with 20,000 copies/well of B2M. The black cluster on the plot represents the negative droplets, the green cluster represents the droplets that are positive for HSV-1 only, the blue cluster represents the droplets that are positive for B2M only, and the orange cluster represents the droplets that are positive for both HSV-1 and B2M targets. B, concentration plot shows merged triplicate wells across an HSV-1 sample dilution series from 0 to 40,000 copies in a constant background of 20,000 copies (66 ng human genomic DNA) of B2M. The blue markers indicate HSV-1 copies/μl and the green markers indicate B2M copies/μl. All error bars are generated by QuantaSoft™ software and represent the 95% confidence interval.
Distinguishing Genomic Variations
Copy number variation (CNV) is a prominent source of interindividual variability in the human genome. CNV has also been associated with cancers, neurological and autoimmune diseases, and adverse drug responses. Reliably identifying such CNVs represents an important capability in cutting-edge research.
The major technical hurdle in copy number (CN) assessment is the ability to discriminate, with statistical confidence, between consecutive CN states. Fundamentally, as CN state increases, the percentage difference in target genomic material between states decreases, making it harder to measure CNV at higher orders. By partitioning the CNV amplification reaction across thousands of droplets, the QX200™ Droplet Digital™ PCR system can resolve high consecutive copy number states (such as five vs. six) with an excellent degree of statistical confidence.
Detection of Copy Number for Myelocytomatosis Oncogene and β-globin
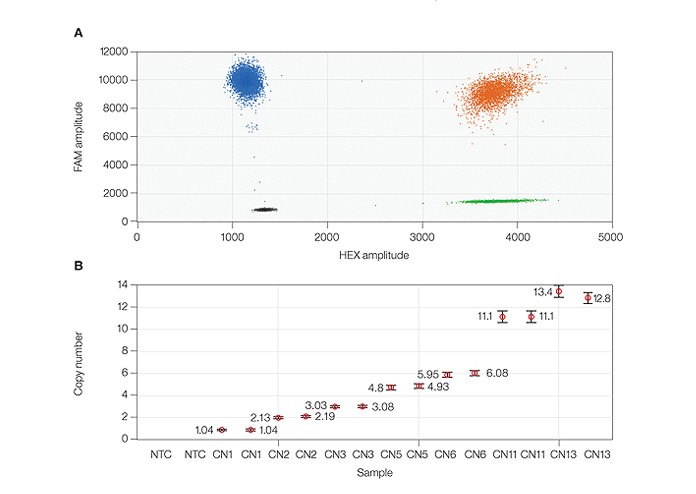
The QX200 system provides superior resolution with copy number calls from CN1 to CN13. A, 2-D fluorescence amplitude plot shows duplicate wells of a CN6 sample duplexed with myelocytomatosis oncogene (MYC) and β-globin (HBB) assays. The black cluster on the plot represents the negative droplets, the green cluster represents the droplets that are positive for MYC only, the blue cluster represents the droplets that are positive for HBB only, and the orange cluster represents the droplets that are positive for both MYC and HBB targets. B, copy number plot shows samples ranging from CN1 to CN13 with precise duplicate well values. All error bars generated by QuantaSoft™ software represent the 95% confidence interval. NTC, no template control.
Next-Generation Sequencing Validation
Accurately quantifying sequencing libraries enables consistent library loading and efficient utilization of next generation sequencing (NGS) platforms. Bio-Rad’s QX200™ Droplet Digital™ PCR system can be easily incorporated into the NGS library preparation workflow to precisely measure and balance sequencing libraries on sequencers. In particular, pre-amplifying in droplets during library preparation ensures better representation of minority species. Along with precise quantification, the data plots generated by the QX200 system are rich with qualitative library information, which is not available with other methodologies. After the NGS run, ddPCR™ can be also used to validate the results obtained by NGS, including genomic alterations such as single nucleotide polymorphisms, mutations, and copy number variations.
Validation for Next-Generation Sequencing
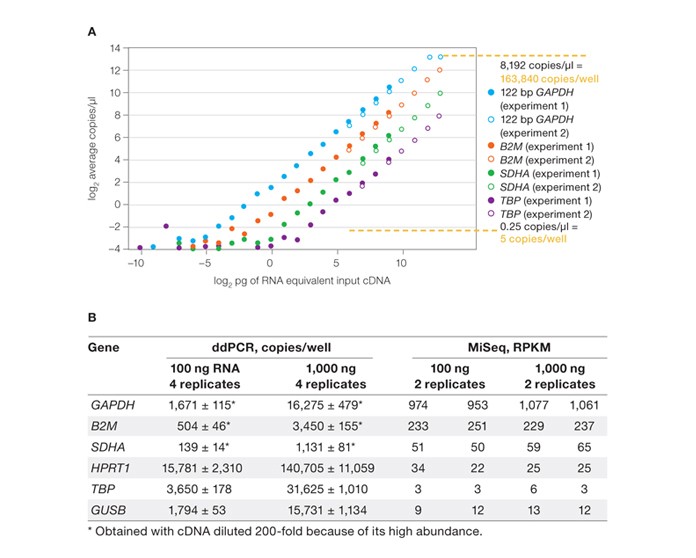
ddPCR provides nearly 1,000-fold more sensitivity and an amplification bias–free approach compared to RNA-Seq for validation. A, concentration plot shows a twofold dilution of cDNA generated using Bio-Rad’s iScript™ advanced cDNA synthesis kit for RT-qPCR. Two independent measurement sets were made: one at a high concentration range and one at a lower range, with four points overlapping. B, table shows comparative results between ddPCR and MiSeq. ddPCR detected thousands of copies per well with low-abundance transcripts (TBP and GUSB genes) using an input of 100 ng total RNA, which demonstrates enhanced sensitivity. The MiSeq sequencer detected only single-digit reads per kilobase per million reads (RPKM) for these transcripts. ddPCR, Droplet Digital PCR.
MiSeq is a trademark of Illumina, Inc.
Gene Expression Studies
Obtaining accurate values for RNA transcripts present in a sample leads to greater understanding of gene expression and regulation. With its high sensitivity, the QX200™ Droplet Digital™ PCR system is especially well-suited for discerning and quantifying rare transcripts, empowering researchers to dive deeper into single cell transcriptomics and gain a better understanding of actual RNA function. With its high precision, the QX200 system can be used to detect small fold changes in expression of a target gene between samples, providing insights into the gene’s physiological implications. The ability to measure absolute values of RNA molecules adds a new level of insight to gene expression studies. The QX200™ ddPCR™ system allows for such measurements without requiring a standard curve.
Gene Expression Analysis of Stem Cell Markers
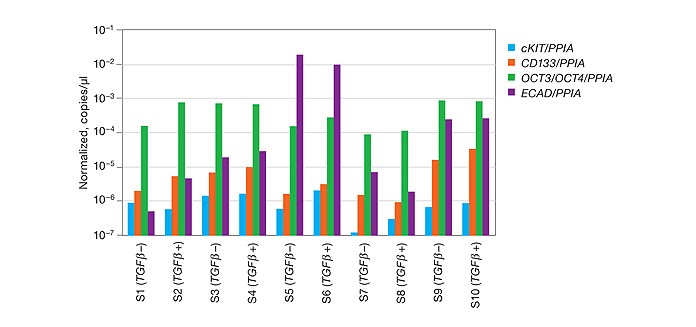
High sensitivity and precision of the QX200 system allow reliable detection of small fold changes with rare transcripts. The TGF-β signaling pathway plays a role in ovarian stem cell regulation, and the addition of TGF-β will increase the number of stem cells in the population, increase the expression of putative stem cell markers, and decrease terminal differentiation markers like E-cadherin (ECAD). Data show expression levels of cKIT, CD133, OCT3/OCT4, and ECAD genes across ten samples with and without TGF-β treatment, normalized to PPIA in this study. Reproducible detection of cDNA copies as low as 0.5–1 target/μl in a 20 μl reaction was achieved. S, sample.
GMO Quantification
Measuring the number of transgene copies present in genetically modified organisms (GMOs) is vital for ensuring that these products meet regulatory and labeling requirements. With its capacity for precise results and high throughput, the QX200™ Droplet Digital™ PCR system offers a reliable platform for analyzing GMO food products at the level required by international standards. By providing absolute quantification of transgene concentrations against reference genes, ddPCR™ offers greater repeatability and cost-effectiveness than qPCR protocols, especially when analyzing low concentrations of target. The QX200 system’s high throughput and ability to run multiplex assays leads to additional cost and reliability advantages over qPCR.
MON810 content measured by ddPCR in five series of seven replicates
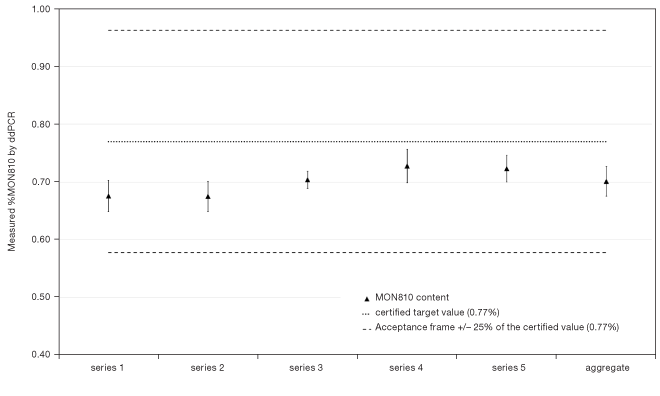
MON810 content measured by ddPCR in five series of seven replicates. The aggregate represents the sum of the five series. The target certified MON810 content (0.77%) is indicated by a dotted line. Acceptance criterion for repeatability is ±25% of the target content (from 0.58% to 0.96%) represented by the dashed lines. Error bars represent the standard deviation between the replicates for each series or in the aggregate.
Morisset D, et al. (2013). Quantitative Analysis of Food and Feed Samples with Droplet Digital PCR. PLoS ONE 8(5): e62583. doi: 10.1371/journal.pone.0062583 CC BY.
Environmental Monitoring
Environmental monitoring for genetic targets, whether from pathogens, invasive species, or other biological activity, requires the ability to detect small target concentrations within complex, non-uniform samples. By creating thousands of PCR reaction chambers in each run, the QX200™ Droplet Digital™ PCR system gives quantitative measurements, even with low concentrations. Because these results are derived by analyzing thousands of reactions run to their endpoints, they are much more tolerant to the presence of PCR inhibitors commonly found in environmental samples that can sometimes alter real-time PCR reaction kinetics and skew qPCR measurements. Getting absolute quantitation rather than relative measures also yields more standardized data when working with multiple monitoring agencies in multiple locations. In addition, using the QX200 system’s multiplexing allows for richer and more meaningful analysis of each environmental sample.
Measuring Two Microbial Targets Simultaneously Using the ddPCR™ System
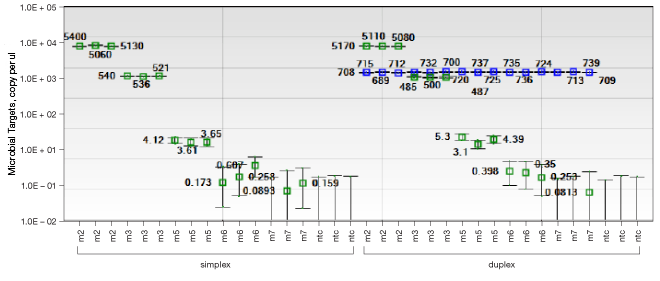
Droplet partitioning by the ddPCR system limits reagent competition and allows for reliable duplex assays in complex environmental samples. Using an environmental water sample, one microbial target was quantified as shown by the green squares in the plot. A duplex assay was performed measuring both target 1 and the target 2 (blue squares), showing that measurement of both targets can be achieved simultaneously without interference.
Ana dyes, Image courtesy of Dr. Yiping Cao from Southern California Coastal Water Research Project.
Legal Notice
The QX200 Droplet Digital PCR system product and/or its use is covered by claims of U.S. patents, and/or pending U.S. and non-U.S. patent applications owned by or under license to Bio-Rad Laboratories, Inc. Purchase of the product includes a limited, non-transferable right under such intellectual property for use of the product for internal research purposes only. No rights are granted for diagnostic uses. No rights are granted for use of the product for commercial applications of any kind, including but not limited to manufacturing, quality control, or commercial services, such as contract services or fee for services. Information concerning a license for such uses can be obtained from Bio-Rad Laboratories. It is the responsibility of the purchaser/end user to acquire any additional intellectual property rights that may be required.

