Droplet Digital PCR (ddPCR) technology is being widely adapted in basic and clinical research applications due to its ability to accurately measure low-abundance targets, rare mutations, allelic variants, targets in complex backgrounds, and to monitor subtle variations in gene expression. Bio-Rad’s Droplet Digital PCR Systems achieve this ability by partitioning a sample into thousands of microscopic droplets prior to amplification. Essentially a PCR reaction occurs within each of these droplets, facilitating accurate and reliable measurements. Droplet Digital PCR technology (described in detail in the Applications & Technology pages of the Bio-Rad website) is used in a wide variety of research applications.
Here are two success stories told by scientists who used Bio-Rad’s QX100™ Droplet Digital PCR System to perform genomic variation analyses.
Detecting Single Nucleotide Variations in induced Pluripotent Stem Cells by Droplet Digital PCR
Research Background
Several recent studies demonstrate that defined transcription factors mediating reprogramming can potentially induce or enlarge mutations in the resulting induced pluripotent stem cells (iPSCs). In our study, DNAs were extracted from several iPSC lines and their matched progenitor somatic cells, then the whole genome was sequenced using a HiSeq 2000 sequencer (Illumina, Inc.). Thousands of single nucleotide variations (SNVs) that occurred in iPSC lines and some high-frequency SNVs, including all the coding ones and randomly selected ones in noncoding regions, were verified by Sequenom genotyping.

PhD Candidate, Gao Shaorong’s Laboratory, National Institute of Biological Sciences
Beijing, China
Application
We used a QX100™ Droplet Digital™ PCR (ddPCR™) system to test whether some observed mutations pre-existed in the starting somatic cells and to further validate the high frequency of SNVs. However, we first performed our experiment using an Applied Biosystems 7500 fast real-time PCR system. We did not trust the results because of the known limited resolution of this instrument. Fortunately, with Bio-Rad’s guidance, we applied genotyping using a QX100 ddPCR system, which allowed us not only to validate the high frequencies of SNVs in iPSC lines, but also to observe the low frequencies of the SNVs in matched progenitor somatic cell lines.
ddPCR Results
Using the QX100 ddPCR system, we further confirmed the high frequency of SNVs in iPSC lines validated by whole genome sequencing and Sequenom genotyping (Figure 1). Moreover, low-frequency SNVs in somatic cells were detected and the cell frequencies were estimated correctly (Figure 2).
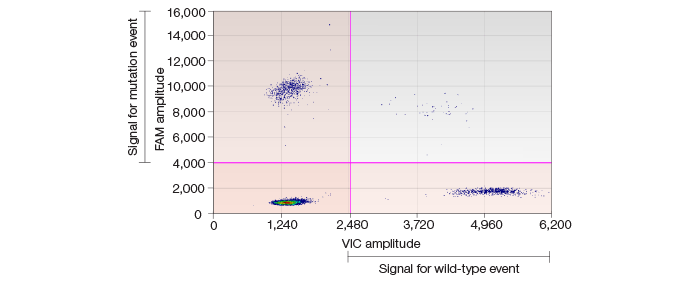
Fig. 1. Scatter plot showing high-frequency SNVs in iPSC lines.
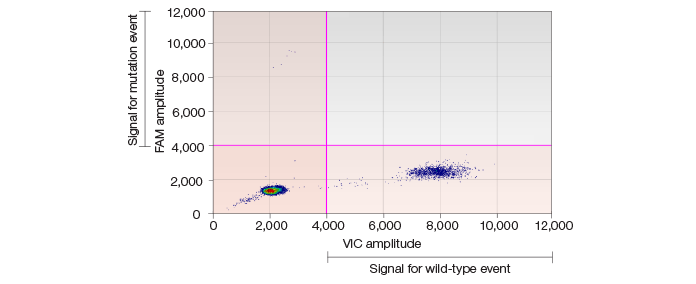
Fig. 2. Scatter plot showing low-frequency SNVs in somatic cells.
In our study, two primary SNV occurrence patterns were demonstrated by the QX100 ddPCR system. One type of SNV had a low frequency only in its corresponding matched progenitor somatic cells and occurred with high frequency in iPSC lines. Another type of SNV had no detectable frequency in parent somatic cells but could be detected in subsequent iPSC lines with high frequency. This latter type is considered a de novo SNV.
Publications
Abyzov A et al. (2012). Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature 492, 438–442.
Hindson BJ et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83, 8604–8610.
Pinheiro LB et al. (2012). Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84, 1003–1011.
For more information, visit bio-rad.com/ddPCRsuccessGao.
Validation of Allele-Specific Expression Predicted by RNA-Seq in Human Brain Specimens
Research Background
The goal of our research is to study genetic mechanisms contributing to the specialization of the human brain in the context of human development and disease occurrence. Technologies used in our laboratory include high-throughput sequencing, microarray, Droplet Digital PCR (ddPCR), various bioinformatics tools, and gene manipulation in model organisms to functionally validate these findings. Allele-specific expression (ASE) is an important factor contributing to human phenotypic variability and the development of complex traits and diseases. We have employed RNA-Seq and SNP chip technology to predict ASE of 16 different brain regions from six healthy adult individuals.

Director, Penn State Hershey Genome Sciences Facility
Assistant Professor of Pharmacology Penn State College of Medicine*
Application
Among candidate alleles with an imbalanced expression from the six healthy individuals, one allele failed to be validated for its genotype by a standard quantitative PCR (qPCR) approach using an inventoried TaqMan assay. The homozygous A/A allele was not successfully resolved due to amplification of the G allele (likely occurring from nonspecific, cross-reactive amplification), generating ambiguous results (Figure 3). Several attempts with different settings yielded the same results. When I was going to move to some other technologies to overcome this problem, the QX100™ Droplet Digital PCR system was launched in our laboratory. We primarily use the ddPCR system to quantify absolute gene expression levels, especially for genes with low expression. I hoped the QX100 ddPCR system could also help assay genotype identification and allelic imbalance with higher precision.
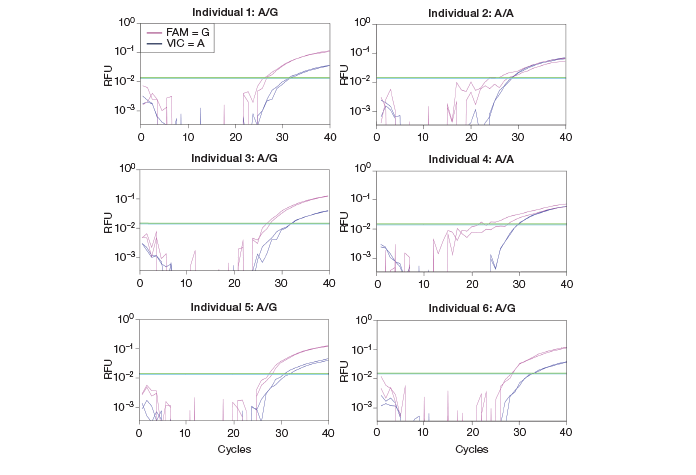
Fig. 3. Real-time PCR amplification curves for a candidate ASE allele. gDNA SNP assays were performed using qPCR. qPCR failed to call the A/A genotype. RFU, relative fluorescence units.
ddPCR Results
As I expected, the QX100 ddPCR system generated a crystal clear, unambiguous result for genotype identification (Figure 4). Results show that the homozygous A/A allele generated nearly zero positive droplets using a G assay (FAM) while generating twice as many droplets with an A assay (VIC) when the data were normalized by their input genomic content (Figure 2). I also saved our precious human genomic DNA (gDNA) by applying ten times less gDNA than the amount used for qPCR. The methods and instrument are robust, producing little variation in results among users once they are trained. I have applied ddPCR to quantify ASE events in cDNA samples obtained from various brain regions of different individuals and I achieved an absolute comparison of gene expression from both alleles (data not shown) without using any standard.
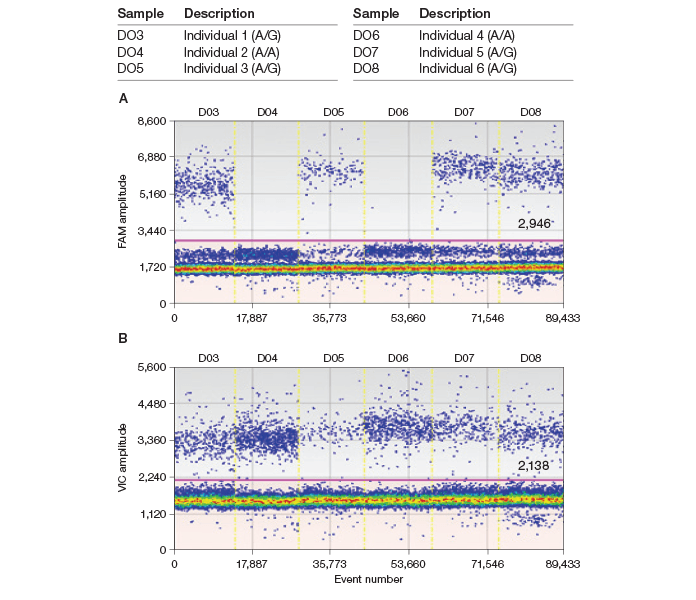
Fig. 4. Droplet Digital PCR validates both A/G and A/A genotypes. gDNA SNP assays were performed with G (A, FAM) and A (B, VIC) assays using ddPCR. A/G and A/A genotypes were successfully validated by ddPCR.
Conclusions
Bio-Rad’s QX100 ddPCR system provides a method for accurately validating genotypes (SNPs). It also allows direct quantification of ASE (allelic imbalance) without the use of a standard, as opposed to standard qPCR that can provide indirect quantification. The costs and time constraints of ddPCR are negligible compared to other known methods, such as the SNaPshot multiplex system, for accurate ASE detection.
Publication
Kang HJ et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489.
For more information, visit bio-rad.com/N/ddPCRSuccessImamura.
HiSeq and Illumina are trademarks of Illumina, Inc. Sequenom is a trademark of Sequenom, Inc.
FAM, SNaPshot, and VIC are trademarks of Applera Corporation. TaqMan is a trademark of Roche Molecular Systems, Inc.
The QX100 Droplet Digital PCR system and/or its use is covered by claims of U.S. patents, and/or pending U.S. and non-U.S. patent applications owned by or under license to Bio-Rad Laboratories, Inc. Purchase of the product includes a limited, non-transferable right under such intellectual property for use of the product for internal research purposes only. No rights are granted for diagnostic uses. No rights are granted for use of the product for commercial applications of any kind, including but not limited to manufacturing, quality control, or commercial services, such as contract services or fee for services. Information concerning a license for such uses can be obtained from Bio-Rad Laboratories. It is the responsibility of the purchaser/end user to acquire any additional intellectual property rights that may be required.

