“Everything is possible with CRISPR. I’m not kidding.” Reacting to highly successful CRISPR gene drive experiments that broke all rules of Mendelian genetics and allowed a mutation to sweep a Drosophila population in a single generation (Hesman Saey 2015), Hugo Bellen, a geneticist at Baylor University, summed up a sentiment shared by many. The speed, low cost, and transferability of CRISPR-Cas9 gene editing to almost any species is redefining what is possible in R&D.
On a monthly basis, more and more new applications for CRISPR are published. From creating complex, multigene disease models and whole-genome knockout screens in less than two months to creating inducible systems and generating Cas9 variants that act as epigenetic regulators, CRISPR is not only accelerating R&D. It appears to be emerging as part of the solution to curbing the ever-increasing cost of drug development and reducing late-stage failure rates.
Faster and cheaper disease models
Within a year of their first successful CRISPR gene drive experiments, scientists at UC San Diego applied the same approach to generate mosquitos that make antimalarial antibodies (Hammond 2016) and several laboratories are now adapting this technology to control the Zika epidemic (Regalado 2016). Similarly rapid advances are being made using CRISPR for the creation of new and improved model systems for drug discovery, target validation, and preclinical trials.
One key to successful drug development is a model system that closely mimics human biology and disease. Traditionally, generating a new disease model was a laborious and expensive task limited to a few genetically tractable species and a handful of cell types with high rates of homologous recombination. To introduce a single desired mutation, mouse embryonic stem cells (mESCs) had to be cultured to identify the tiny fraction of correctly edited cells. Those cells then had to be introduced into early-stage embryos, resulting in mosaic mice that were crossed to generate mice with the desired mutation. This process could easily consume two years and cost upwards of US $20,000, and success rates ranged from only 50 to 55% (Cohen 2016). Faced with these facts scientists often had to settle for less than optimal even though they could easily envision the mutations required to generate a better disease model.
CRISPR reduces the number of crosses required to generate disease models. A mouse model, for example, can now be generated in as little as two months, at a fraction of the cost, with success rates of 85 to 95% (Qin and Wang 2016, Cohen 2016). The CRISPR system can be injected directly into mouse embryos, bypassing the need for mESC culture and the breeding of mosaic mice. And it requires only a simple guide RNA that can be ordered for less than US $30. By generating double stranded breaks in DNA at sites complementary to the guide RNA sequence, the CRISPR system allows highly efficient gene editing (Figure 1) (Quinlan 2016). Moreover, CRISPR also works in adult somatic cells, greatly expanding the types of cells that can be genetically manipulated. This allows genome editing directly in adult animals, eliminating the time it takes for embryos to mature; by injecting the CRISPR system into adult mice scientists have created various tumor models (Hou et al. 2015, Weber et al. 2015) and corrected disease-causing mutations associated with hemophilia and hereditary tyrosinemia (Guan et al. 2016, Yin et al. 2014).
Thousands of mutations in a single experiment
CRISPR has not only made mouse model generation fast and cost effective, it is allowing us to conduct highly efficient genome-wide screens in a matter of weeks (Shalem et al. 2015). CRISPR’s simple targeting method allows pooling of guide RNAs that target Cas9 to multiple genomic loci. Using pooled libraries that target more than 20,000 protein coding genes in human and mouse cell lines a range of actionable factors have been identified, including ones that confer resistance to chemotherapeutic agents (Shalem et al. 2014) and Zika and dengue virus (Savidis et al. 2016). Such screens are unthinkable using alternate genome editing systems such as zinc fingers, for which the price of a genome-wide library is estimated to be anywhere from US $15 to 50 million (Moore 2015). Similar screens have been carried out using RNAi, but these experiments are plagued by off-target effects, poor reproducibility, and the fact that knockdown is short lived and often incomplete and variable (Moore 2015).
More clinically relevant model systems
CRISPR also has the potential to address the problem that many promising drug candidates currently fail in the transition from preclinical testing to early-stage clinical trials, when a drug moves from a model system to human patients. Inadequate model systems are at least partially to blame for the high failure rate. Unlike traditional gene editing techniques that were limited to a small number of species, CRISPR has been used successfully in a wide range of species. This has resulted in a vast expansion of available model systems tailored to the study of specific diseases. Ferrets, for example, can now be used as influenza transmission models, taking advantage of the fact that unlike mice, ferrets sneeze when infected (Reardon 2016). Macaque models have already been used to recapitulate some of the symptoms of autism (Liu et al. 2016).
CRISPR also allows R&D to overcome the obstacle that many human diseases are not monogenic and single-mutation disease models do not account for all aspects of a given disorder. By multiplexing guide RNAs, scientists at eGenesis were able to edit the pig genome in 62 places in a single experiment (Reardon 2015). Not only was the pig genome uneditable before CRISPR, generating 62 mutations would have been impossible over the course of a scientist’s entire career. Similar approaches have been used to create complex in vivo models, including ones for liver and lung cancer as well as glioblastoma (Xue et al. 2014, Sánchez-Rivera et al. 2014, Toledo et al. 2015) that will be able to better mimic patient biology.
The ease of gene editing with CRISPR is also accelerating the adoption of powerful but previously costly and technically challenging models, such as mice with humanized immune systems and paired isogenic cell lines. Humanized mice are a key to realizing a promising new approach to cancer therapy, immuno-oncology, which aims to use a patient’s own immune system to eliminate cancer cells. Humanized mice are crucial to developing such therapies because the behavior of transplanted patient derived tumors and the humanized mouse’ immune system can be monitored in response to different treatments.
Paired isogenic cell lines, on the other hand, allow creation of highly controlled and in some cases patient-specific disease models. These cell lines are genetically and epigenetically identical with the exception of specific mutations created using CRISPR. Thus they allow us to confidently assign phenotypes to specific genotypes and to screen for drug candidates that show genotype-specific toxicity (Haagensen et al. 2016).
Beyond editing – gene regulation, epigenetics, and less costly dosage studies
Some of the most promising CRISPR applications take advantage of noncutting Cas9 (dCas9) enzyme. By fusing dCas9 to a repressor domain guide, RNAs can target the CRISPR system to downregulate transcription of target genes in human cells up to 99% (Gilbert et al. 2014). Similarly, fusing dCas9 to an activator allows selective activation of genes for gain-of-function screens (Gilbert et al. 2014). An additional layer of control can be added to these experiments by rendering them photo- or chemi-inducible to allow age/developmental stage–dependent studies as well as manipulation of genes that are embryonic lethal. Some systems even allow spatial control of gene expression to provide even more exquisite control of gene activation (Polstein and Gersbach 2015).
dCas9 has also emerged as a robust tool for the study of epigenetics. A growing number of human diseases and conditions, including heart disease, various cancers, and autism have been linked to epigenetic dysregulation, yet the contribution of epigenetics to disease has been notoriously difficult to study. The dCas9-p300 system enables fast and targeted acetylation of histones by using guide RNAs to target dCas9 fused to the human acetyltransferase p300 (Hilton et al. 2015). A separate system allows targeted demethylation of DNA and has been used in neurons as well as mice to demethylate and thus activate promoter sequences (Xu et al. 2016). Together these systems provide a formidable tool for understanding how epigenetics can drive disease.
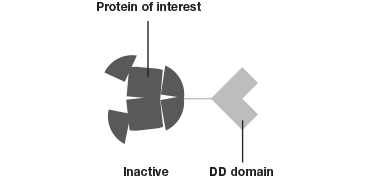
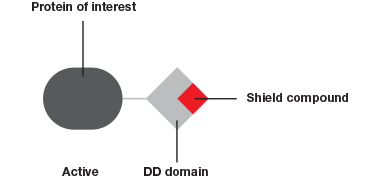
The Degron-KI approach (Figure 1), on the other hand, enables the extent of downregulation to be modulated with precise control. This system has allowed Novartis and the Broad Institute to mimic dosage studies without needing to spend months or even years to identify a small molecule to target the gene of interest (Zhou et al. 2015). A tag added to genes of interest leads to degradation of the resulting protein in the absence of a shield compound. By varying the concentration of shield compound this model system permits dosage studies at a fraction of what they used to cost.
The Many Uses of CRISPR
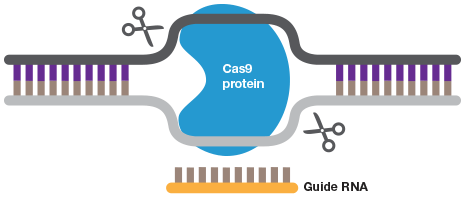
A short guide RNA can be used to target Cas9 to a sequence of interest. Cas9 introduces a double strand DNA break to allow highly efficient gene editing.
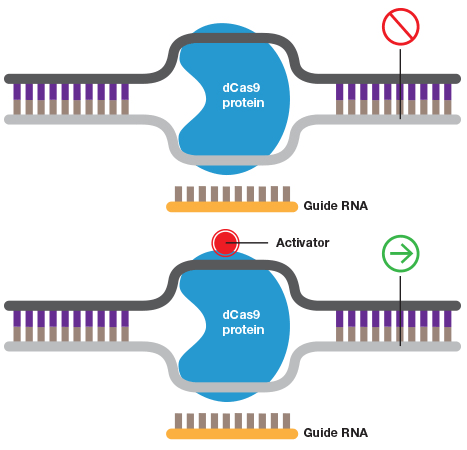
A guide RNA can also be used to target catalytically inactive Cas9 (dCas9) to a promoter to inhibit transcription by blocking transcriptional machinery from binding or to activate transcription when dCas9 is fused to an activator.
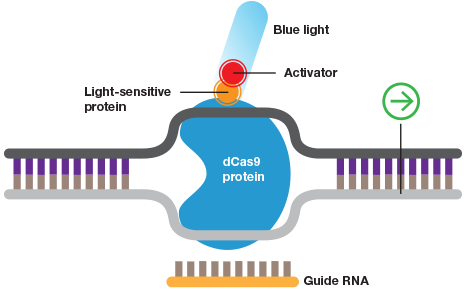
Wild-type Cas9 or dCas9 can be fused to switches that are controlled by light or a specific chemical.
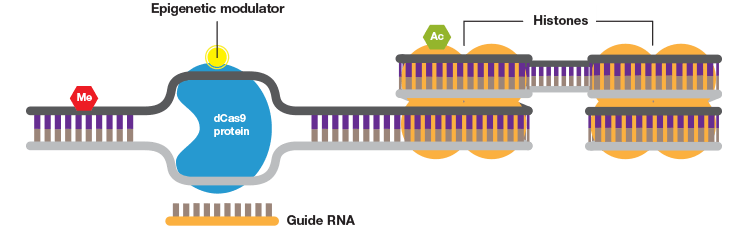
When fused to a DNA methylase or histone acetylase the CRISPR system can be used to study the effects of specific DNA and histone modifications.
The Degron-KI approach uses CRISPR-Cas9 to add a destabilization domain (DD) to proteins of interest. By titrating the shield compound protein levels can be modulated.
Everything is possible
With each new CRISPR-Cas9 application or disease model Hugo Bellen’s words seem less hyperbolic. Month by month more biotechnology and pharmaceutical companies are acknowledging the promise of CRISPR by incorporating this technology into their R&D efforts. Industry leader Jackson Laboratories, for example, recently joined forces with CRISPR start-up Caribou to begin using CRISPR to generate its customized mouse models faster and at lower cost, and both Astra-Zeneca and Novartis have entered into partnerships to use CRISPR for drug screening and validation. CRISPR is undoubtedly redefining what is possible for R&D, and the growing toolkit that exploits CRISPR’s targeting abilities to modify DNA sequences makes Bellen’s words ring truer than ever – the limits of what is possible using CRISPR-Cas9 are increasingly difficult to imagine.
References
Cohen J (2016). ‘Any idiot can do it.’ Genome editor CRISPR could put mutant mice in everyone’s reach. http://www.sciencemag.org/news/2016/11/any-idiot-can-do-it-genome-editor-crispr-could-put-mutant-mice-everyones-reach, accessed December 12, 2016.
Gilbert LA et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661.
Guan Y et al. (2016). CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse.EMBO Mol Med 8, 477–488.
Haagensen EJ et al. (2016). Pre-clinical use of isogenic cell lines and tumours in vitro and in vivo for predictive biomarker discovery; impact of KRAS and PI3KCA mutation status on MEK inhibitor activity is model dependent. Eur J Cancer 56, 69–76.
Hammond A et al. (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol 34, 78–83.
Hesman Saey T (2015). Gene drives spread their wings. https://www.sciencenews.org/article/gene-drives-spread-their-wings, accessed December 12, 2016.
Hilton IB et al. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33, 510–517.
Liu Z et al (2016). Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 530, 98–102.
Moore JD (2015). The impact of CRISPR-Cas0 on target identification and validation. Drug Discov Today 20, 450–457.
Mou H et al. (2015). Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med 7, 53.
Polstein LR and Gersbach CA (2015). A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11, 198–200.
Qin W and Wang H (2016). Generating mouse models using CRISPR/Cas9. http://blog.addgene.org/generating-mouse-models-using-crispr/cas9, accessed December 12, 2016.
Quinlan A (2016). From megaTALs to CRISPR: The many ways to edit a gene. http://www.bioradiations.com/from-megatals-to-crispr-the-many-ways-to-edit-a-gene/, Accessed December 12, 2016.
Reardon S (2015). Gene-editing record smashed in pigs. http://www.nature.com/news/gene-editing-record-smashed-in-pigs-1.18525, accessed December 12, 2016.
Reardon S (2016). Welcome to the CRISPR zoo. Nature 531, 160–163.
Regalado A (2016). We have the technology to destroy all Zika Mosquitos. https://www.technologyreview.com/s/600689/we-have-the-technology-to-destroy-all-Zika-mosquitoes/, accessed December 12, 2016.
Sánchez-Rivera FJ et al. (2014). Rapid modeling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431.
Savidis G et al. (2016). Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep 16, 232–246.
Shalem O et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87.
Shalem O et al. (2015). High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16, 299–311.
Toledo CM et al. (2015). Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in gliablastoma stem-like cells. Cell Rep 13, 2,425–2,439.
Weber J et al. (2015). CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. PNAS 112, 13,982–13,987.
Xu X et al. (2016). A CRISPR-based approach for targeted DNA methylation. Cell Discov 2:16009.
Xue W et al. (2014). CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384.
Yin H et al. (2014). Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32, 551–553.
Zhou Q et al. (2015). A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res 15, 1,949–1,958.

