Abstract
Characterization of gene expression pattern changes can inform our understanding of disease progression as well as the development of better prognostic tools and treatment options. These studies, however, require analysis of large numbers of transcripts and patient samples, which leads to time-consuming and cumbersome workflows. Here we describe a semi-automated quantitative PCR (qPCR) approach, using PrimePCR™ Panels and the CFX Automation System II Plate Handler, that reduces setup time and minimizes the amount of time spent processing qPCR plates.
Figures
|
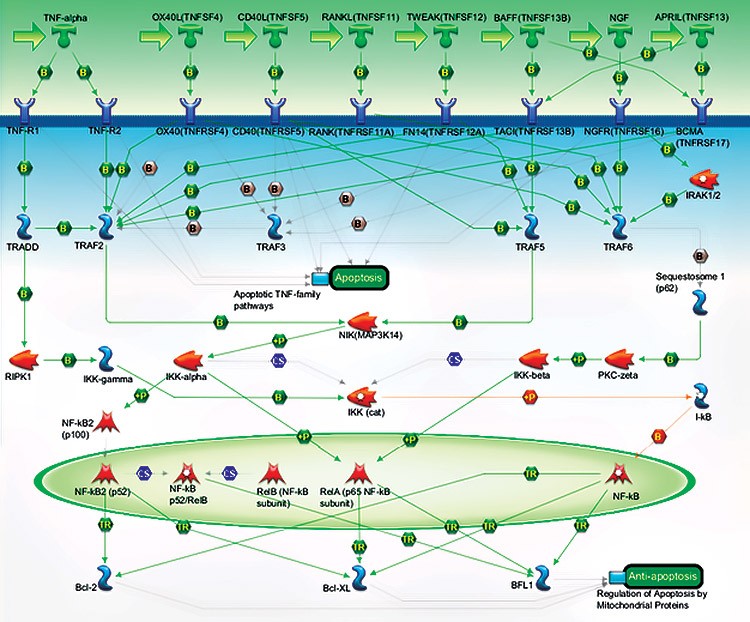
Pathway Map of TNFs/NF-kB/Bcl-2 Figure 1. Pathway map with relationship of targets in the PrimePCR TNFs/NF-kB/Bcl-2 Panel. |
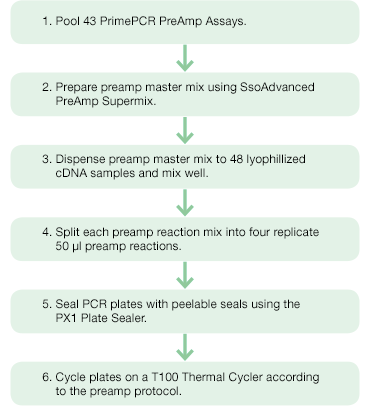
Figure 2. Target-specific preamplification of cDNA of 43 genes of interest using PrimePCR PreAmp Assays and SsoAdvanced PreAmp Supermix. |
|
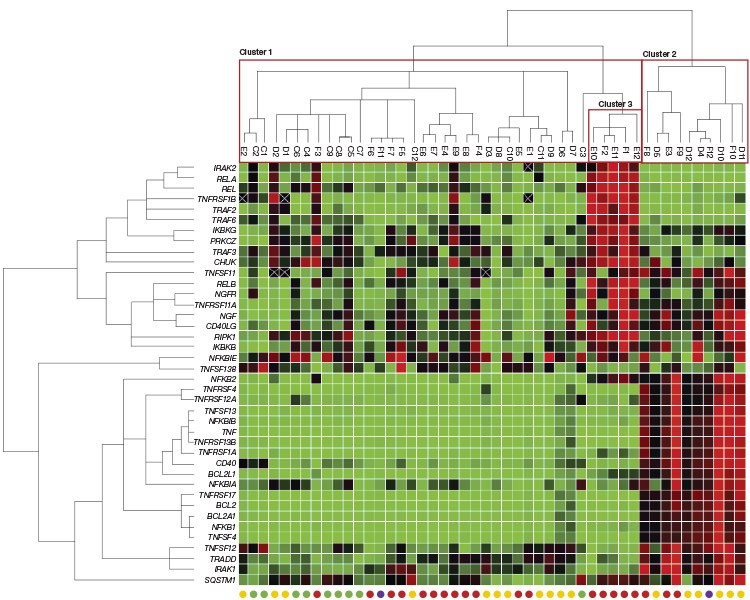
Figure 3. Gene expression patterns in normal and cancerous prostate tissues. |
Introduction
The ability to assess the differential expression of many genes in a large number of patient samples simultaneously plays an important role in understanding complex diseases. Characterizing changes in gene expression patterns during disease progression can aid in the development of better disease prognostics and will help drive toward a more personalized approach to treatment regimens.
We set out to develop a semi-automated workflow that would allow us to more efficiently examine how the expression of genes in the anti-apoptotic TNFs/NF-kB/Bcl-2 pathway varied across samples obtained from patients with various stages of prostate cancer. This pathway was chosen because the evasion of apoptosis plays a fundamental role in tumorigenesis (Wong 2011) and its role in prostate cancer progression is well documented (McDonnell et al. 1992).
Since the need to analyze many genes across multiple patient samples quickly increased the complexity of qPCR setup, the CFX Automation System II, a plate handling system that automates plate loading and data collection on CFX Real-Time PCR Detection Systems, was used in conjunction with PrimePCR Panels to streamline the qPCR workflow. Gene expression profiles were analyzed in prostate samples from 39 patients with prostate cancer and nine healthy adults using the PrimePCR Anti-Apoptotic TNFs/NF-kB/Bcl-2 Pathway H384 (human, 384-well) Panel (Figure 1). This panel provides lyophilized primer sets for the analysis of 40 genes of interest in the TNFs/NF-kB/Bcl-2 pathway along with three reference genes (HPRT1, GAPDH, and TBP) for analysis of up to eight samples. Since only small amounts of each sample were available (2–3 ng cDNA), preamplification was used prior to qPCR to ensure that enough sample was available for the analysis of all the genes of interest. This semi-automated workflow translated into an approximately 50% reduction in hands-on time and required approximately 78% less total time spent in or near the laboratory to conduct the experiment.
A. TNFs/NF-kB/Bcl-2 Pathway Map
B. Target List for PrimePCR Anti-Apoptotic TNFs/NF-kB/Bcl-2 Pathway
| CL2 | IRAK2 | RELB | TNFRSF4 |
| BCL2A1 | NFKB1 | RIPK1 | TNFSF11 |
| BCL2L1 | NFKB2 | SQSTM1 | TNFSF12 |
| CD40 | NFKBIA | TBP | TNFSF13 |
| CD40LG | NFKBIB | TNF | TNFSF13B |
| CHUK | NFKBIE | TNFRSF11A | TNFSF4 |
| GAPDH | NGF | TNFRSF12A | TRADD |
| HPRT1 | NGFR | TNFRSF13B | TRAF2 |
| IKBKB | PRKCZ | TNFRSF17 | TRAF3 |
| IKBKG | REL | TNFRSF1A | TRAF6 |
| IRAK1 | RELA | TNFRSF1B |
Fig. 1. Gene targets assayed using the PrimePCR Anti-Apoptotic TNFs/ NF-kB/Bcl-2 Pathway Panel. A, pathway map showing the relationship of targets included in the PrimePCR TNFs/NF-kB/Bcl-2 Panel; B, list of targets included in the PrimePCR Anti-Apoptotic TNFs/NF-kB/Bcl-2 Pathway Panel.
Methods
Sample Source
cDNA from nine healthy and 39 cancerous prostates were obtained from OriGene Technologies.
cDNA Preamplification
To obtain sufficient amounts of cDNA, preamplification was performed using SsoAdvanced™ PreAmp Supermix and PrimePCR PreAmp Assays (Figure 2). The 43 PrimePCR PreAmp Assays were pooled and diluted 100-fold with PCR-grade water to create the preamplification assay pool. A master mix sufficient for 192 reactions (48 samples with four replicates each) was prepared such that each 50 µl preamplification reaction contained 25 µl SsoAdvanced PreAmp Supermix, 5 µl preamplification assay pool, and 20 µl nuclease-free water. Two hundred microliters of master mix were dispensed into each lyophilized cDNA sample and mixed well. The resulting reaction mixes were split into four replicate 50 µl preamplification reactions in 96-well PCR plates and sealed with Peelable Foil Heat Seals using the PX1™ PCR Plate Sealer. The reactions were cycled in a T100™ Thermal Cycler according to the SsoAdvanced PreAmp Supermix protocol using 12 preamplification cycles (95°C, 3 min; 12 cycles of [95°C, 15 sec; 58°C, 4 min]; 4°C hold ) for a 4,096-fold theoretical target amplification. Preamplified cDNA was then diluted fivefold using TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8) prior to its use in the qPCR reaction.
Fig.2. Preamplification workflow. PrimePCR PreAmp Assays and SsoAdvanced PreAmp Supermix were used for target-specific preamplification of cDNA of 43 genes of interest.
Quantitative PCR
After preamplification, a qPCR master mix was prepared for each of the 192 preamplified samples such that a single 10 µl reaction contained 5 µl SsoAdvanced™ Universal SYBR® Green Supermix, 1 µl preamplified cDNA, and 4 µl nuclease-free water. SsoAdvanced™ Universal SYBR® Green Supermix was chosen for its superior stability at room temperature. Ten microliters of the master mix were dispensed into each well of the PrimePCR Anti-Apoptotic TNFs/NF-kB/Bcl-2 Pathway H384 Plates. PrimePCR Plates were sealed with Optically Clear Heat Seals using the PX1 PCR Plate Sealer. Sealed qPCR plates were then centrifuged at 4,000 x g for 4 min and loaded into the CFX Automation System II.
The CFX Automation System II was configured with two CFX384™ Real-Time PCR Detection Systems for this experiment. The PrimePCR run file containing gene targets and control assay information for the PrimePCR Anti-Apoptotic TNFs/NF-kB/Bcl-2 Pathway H384 Panel was downloaded from the PrimePCR website and imported into the automation system software. Reactions were cycled using a standard protocol (95°C, 2 min; 40 cycles of [95°C, 5 sec; 60°C, 30 sec]; melt curve analysis: 65–95°C, 0.5°C increments, 5 sec hold). The automation system was started and proceeded to run the 24 panels over an 18.2 hour period.
Data Analysis
Data analysis was conducted using CFX Manager™ Software. Each file was annotated with its corresponding sample name and the individual data files were imported into a CFX Manager gene study file, where the results across all 24 PrimePCR Plates were compiled.
The clustergram module in CFX Manager Software was used to analyze the anti-apoptotic gene expression profiles for this set of prostate tissue samples. The clustergram assembles samples and/or gene targets into hierarchies based on similarities in gene expression and is appropriate for use in relative quantification (ΔCq) experiments consisting of unpaired samples.
Fig. 3. Gene expression patterns in normal and cancerous prostate tissues. Red squares indicate upregulated genes, green squares downregulated genes, and black squares genes that showed no change in expression. An X indicates that no transcript was detected. Circles at the bottom of each column indicate cancer status of each prostate tissue sample: normal (●); stage II (●); stage III (●); stage IV (●).
Results
Inspection of the qPCR results clustered by sample reveals that two primary clusters of samples exist: one consisting of normal and cancerous prostate tissues (Cluster 1 in Figure 3) and another that is dominated by stage II–IV prostate cancer samples (Cluster 2 in Figure 3). This second cluster shows the upregulation of BCL2 and associated genes, indicating the inhibition of apoptosis mediated by the Bcl-2 pathway (Cory et al. 2003). The clustering analysis also revealed a secondary expression profile (Cluster 3 in Figure 3) specific to a subset of the stage III prostate cancer samples within Cluster 1. This expression profile is marked by the upregulation of REL, RELA, TRAF2, and RIPK1, likely indicating activation of NF-kB and, ultimately, inhibition of apoptosis mediated by XIAP, c-IAP1, and c-IAP2 (de Almagro and Vucic 2012). While not performed here, expression changes in this pathway could be further explored using the PrimePCR Anti-Apoptotic TNFs/NF-kB/IAP Pathway Panel.
Conclusions
As the requirement for qPCR throughput in a laboratory grows, the complexity of and time requirement for qPCR setup rise proportionally. In this study, several key products were used to develop a semi-automated qPCR workflow that reduces the experimental complexity and setup time (Table 1). PrimePCR Panels simplified reaction setup because the lyophilized primer sets are already dispensed in the qPCR plates, thus removing a liquid handling step during the qPCR setup and reducing total hands-on time. By using the CFX Automation System II Plate Handler with two CFX384 Real-Time PCR Detection Systems, we were able to collect qPCR data for all twenty-four 384-well plates over a period of 18.2 hours (506 data points per hour) while leaving the instrument unattended. Compared to a conventional workflow, in which each qPCR plate must be loaded into the real-time PCR instrument manually, this amounted to approximately 78% less time spent in or near the laboratory over the course of the experiment (Table 1). Additionally, the use of SsoAdvanced™ Universal SYBR® Green Supermix, which is stable at room temperature for up to 48 hours, ensured that the assembled qPCR reactions remained stable at room temperature until they were processed. This new workflow enabled us to obtain 8,256 data points across 48 patient samples in 29.3 hours, approximately 5 hours faster than with a conventional workflow, and with approximately 50% less hands-on time. This workflow can easily be converted into a fully automated process through the addition of a liquid handling system, which would permit even higher throughput analysis of gene expression patterns associated with disease.
Table 1. Approximate time savings using the semi-automated PrimePCR workflow.
| Fully Manual qPCR Setup | qPCR Setup with PrimePCR Assays and the CFX Automation System II | |
| Preamplification of 48 cDNA samples with 4 replicates | 2.1 hr (1 hr hands-on) |
2.1 hr (1 hr hands-on) |
| qPCR setup for 192 samples and 43 gene targets | 7 hr (all hands-on) |
4 hr (all hands-on) |
| qPCR data collection | 20.2 hr (2 hr* hands-on) |
18.2 hr (5 min hands-on) |
| Total time to results | 29.3 hr | 24.3 hr (17.1% reduction) |
| Total estimated hands-on time | 10 hr | 5 hr (50% reduction) |
| Total time required in laboratory** | 28.2 hr | 6.2 hr (78% reduction) |
* 5 min per plate
** This reflects the amount of time required to perform preamplification, qPCR setup, plate loading, and initiation of qPCR cycling. For the fully manual setup, this duration includes the cycling time for all but the last pair of plates, because each plate has to be loaded manually.
Visit bio-rad.com/qPCRsemiautomated for more information.
References
Cory S et al. (2003). The Bcl-2 family: Roles in cell survival and oncogenesis. Oncogene 22, 8,590–8,607.
de Almagro MC and Vucic D (2012). The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp Oncol 34, 200–211.
McDonnell TJ et al. (1992). Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res 52, 6,940–6,944.
Wong RS (2011). Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res 30, 87.
Related Information
Bureeva S et al. (2012). PrimePCR™ pathway analysis: Pathway curation and real-time PCR panel design strategy. Bio-Rad Bulletin 6263.
Hellemans J et al. (2012). PrimePCR™ Assays: Meeting the MIQE guidelines by full wet-lab validation. Bio-Rad Bulletin 6262.
OriGene is a trademark of OriGene Technologies, Inc. SYBR is a trademark of Life Technologies Corporation. Bio-Rad Laboratories, Inc. is licensed by Life Technologies Corporation to sell reagents containing SYBR Green I for use in real-time PCR, for research purposes only.
Bio-Rad’s thermal cyclers and real-time thermal cyclers are covered by one or more of the following U.S. patents or their foreign counterparts owned by Eppendorf AG: U.S. Patent Numbers 6,767,512 and 7,074,367.
The use of SsoAdvanced Supermixes and PrimePCR PreAmp Assays is covered by one or more of the following U.S. patents and corresponding patent claims outside the U.S.: 5,804,375; 5,994,056; and 6,171,785. The purchase of these products includes a limited, non-transferable immunity from suit under the foregoing patent claims for using only this amount of product for the purchaser’s own internal research. No right under any other patent claim and no right to perform commercial services of any kind, including without limitation reporting the results of purchaser’s activities for a fee or other commercial consideration, are conveyed expressly, by implication, or by estoppel. These products are for research use only. Diagnostic uses under Roche patents require a separate license from Roche. Further information on purchasing licenses may be obtained from the Director of Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.